Difference between revisions of "Part:BBa K2047002"
Lindsay.Li (Talk | contribs) |
|||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| − | <partinfo> | + | <partinfo>BBa_K2047002 short</partinfo> |
| − | + | <h3>Introduction</h3> | |
<p style="font-size:16px;font-family:'Calibri'">Inspired by Xu’s work and based on keasling’s work, we designed a series of stem-loops with different free energy for further use as basic regulatory parts. To measure the regulation effect of stem-loop, we constructed the dual-fluorescent reporter system (GFP and mCherry) to test the regulatory effect of various stem-loops.</p> | <p style="font-size:16px;font-family:'Calibri'">Inspired by Xu’s work and based on keasling’s work, we designed a series of stem-loops with different free energy for further use as basic regulatory parts. To measure the regulation effect of stem-loop, we constructed the dual-fluorescent reporter system (GFP and mCherry) to test the regulatory effect of various stem-loops.</p> | ||
<p style="font-size:16px;font-family:'Calibri'">The operon is transcribed by its sole promoter and the primary transcript is cleaved into several secondary transcripts by RNase E, a single-stranded, nonspecific endonuclease with preference for cleaving A/U-rich sequence. However, the stability of these secondary transcripts against exonuclease degradation from the 3’ end varied due to their distinct terminal structure. When stem-loops inserted in the 3'end of the upstream gene, it protects its mRNA against the cleavage of exonuclease, increasing the ratio of abundance of the first gene product relative to that of the second gene product. Furthermore, the lower free energy of stem-loops are, the more stable the secondary transcripts of the upstream are, tuning the expression of multiple genes.</p> | <p style="font-size:16px;font-family:'Calibri'">The operon is transcribed by its sole promoter and the primary transcript is cleaved into several secondary transcripts by RNase E, a single-stranded, nonspecific endonuclease with preference for cleaving A/U-rich sequence. However, the stability of these secondary transcripts against exonuclease degradation from the 3’ end varied due to their distinct terminal structure. When stem-loops inserted in the 3'end of the upstream gene, it protects its mRNA against the cleavage of exonuclease, increasing the ratio of abundance of the first gene product relative to that of the second gene product. Furthermore, the lower free energy of stem-loops are, the more stable the secondary transcripts of the upstream are, tuning the expression of multiple genes.</p> | ||
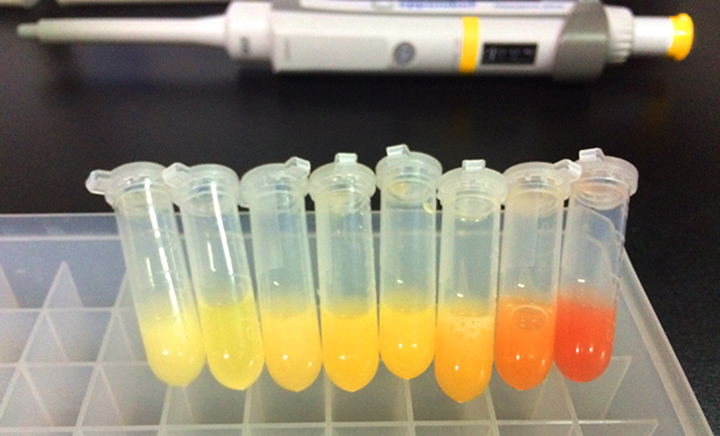
[[File:T--OUC-China--part-expression.jpg|center|thumb|400px|Figure 1 shows the expression of fluorescence protein effected by stem-loops with different folding free energy. It’s clearly that the difference is significant. Then we measured the quantitative expression on two levels: the transcriptional level and the translational level.]] | [[File:T--OUC-China--part-expression.jpg|center|thumb|400px|Figure 1 shows the expression of fluorescence protein effected by stem-loops with different folding free energy. It’s clearly that the difference is significant. Then we measured the quantitative expression on two levels: the transcriptional level and the translational level.]] | ||
[[File:T--OUC-China--part-all.png|center|thumb|400px|Figure.2 shows the relative expression level of the dual-fluorescence reporter system we designed. The ratio of GFP and mCherry has significant difference both on the transcriptional level and the translational level with different stem-loops.]] | [[File:T--OUC-China--part-all.png|center|thumb|400px|Figure.2 shows the relative expression level of the dual-fluorescence reporter system we designed. The ratio of GFP and mCherry has significant difference both on the transcriptional level and the translational level with different stem-loops.]] | ||
| − | <h3>Description</h3 | + | <h3>Description</h3> |
<p style="font-size:16px;font-family:'Calibri'">We designed a series of stem-loops followed by a RNase site with various free energy measured by Mfold, which can be used as basic regulatory elements.</p> | <p style="font-size:16px;font-family:'Calibri'">We designed a series of stem-loops followed by a RNase site with various free energy measured by Mfold, which can be used as basic regulatory elements.</p> | ||
<p style="font-size:16px;font-family:'Calibri'">This part encodes stem-loop of -14.9 kcal/mol that designed by other researchers and a RNase site downstream. The effect of transcript protection we measured of the stem-loop and RNase is as follows:</p> | <p style="font-size:16px;font-family:'Calibri'">This part encodes stem-loop of -14.9 kcal/mol that designed by other researchers and a RNase site downstream. The effect of transcript protection we measured of the stem-loop and RNase is as follows:</p> | ||
| − | [[File:T--OUC-China--basic-002. | + | [[File:T--OUC-China--basic-002.jpg|center|thumb|400px|Figure 3 Shows the structure of the stem-loop of this part with the folding free energy of -14.9 kcal/mol.]] |
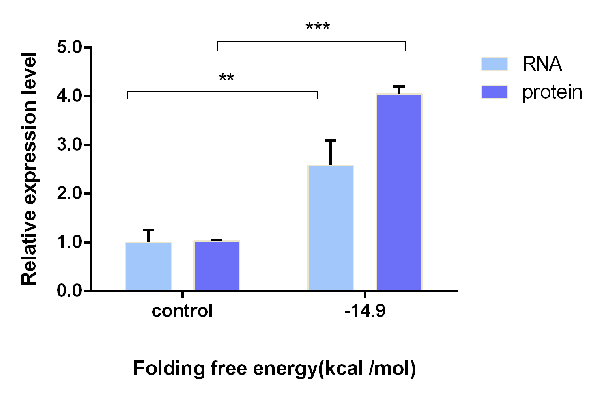
| − | [[File:T--OUC-China--composite-102.png|center|thumb|400px|Figure 4 Shows the relative expression on RNA and protein level with stem-loop of -14.9 kcal/mol (measured by Mfold) contrast to the control group with no stem-loop. The result is the ratio of upstream gfp to downstream mCherry. Error bars indicate s.d. of mean of experiments in triplicate. | + | [[File:T--OUC-China--composite-102.png|center|thumb|400px|Figure 4 Shows the relative expression on RNA and protein level with stem-loop of -14.9 kcal/mol (measured by Mfold) contrast to the control group with no stem-loop. The result is the ratio of upstream gfp to downstream mCherry. Error bars indicate s.d. of mean of experiments in triplicate.(***P value<0.0001, **P value<0.01)]] |
| Line 22: | Line 22: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | <!-- --> | ||
| + | <span class='h3bb'>Sequence and Features</span> | ||
| + | <partinfo>BBa_K2047002 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | <!-- Uncomment this to enable Functional Parameter display | ||
| + | ===Functional Parameters=== | ||
| + | <partinfo>BBa_K2047002 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
Latest revision as of 07:53, 21 October 2016
Stem-loop with free energy of -14.9 kcal/mol measured by Mfold
Introduction
Inspired by Xu’s work and based on keasling’s work, we designed a series of stem-loops with different free energy for further use as basic regulatory parts. To measure the regulation effect of stem-loop, we constructed the dual-fluorescent reporter system (GFP and mCherry) to test the regulatory effect of various stem-loops.
The operon is transcribed by its sole promoter and the primary transcript is cleaved into several secondary transcripts by RNase E, a single-stranded, nonspecific endonuclease with preference for cleaving A/U-rich sequence. However, the stability of these secondary transcripts against exonuclease degradation from the 3’ end varied due to their distinct terminal structure. When stem-loops inserted in the 3'end of the upstream gene, it protects its mRNA against the cleavage of exonuclease, increasing the ratio of abundance of the first gene product relative to that of the second gene product. Furthermore, the lower free energy of stem-loops are, the more stable the secondary transcripts of the upstream are, tuning the expression of multiple genes.
Description
We designed a series of stem-loops followed by a RNase site with various free energy measured by Mfold, which can be used as basic regulatory elements.
This part encodes stem-loop of -14.9 kcal/mol that designed by other researchers and a RNase site downstream. The effect of transcript protection we measured of the stem-loop and RNase is as follows:
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 43
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]