Difference between revisions of "Part:BBa K1980010"
(→Absorbance Assay) |
|||
| (19 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
==Description== | ==Description== | ||
| − | <p>This part contains the bacterial copper chelator Csp1 (Copper Storage Protein 1) with a C-terminal sfGFP tag (connected with short hydrophilic flexible linker) behind the pCopA promoter with | + | <p>This part contains the bacterial copper chelator Csp1 (Copper Storage Protein 1) with a C-terminal sfGFP tag (connected with short hydrophilic flexible linker) behind the pCopA promoter with divergently expressed CueR (a bacterial copper sensitive repressor/activator).</p> |
| − | |||
<!-- --> | <!-- --> | ||
<!-- --> | <!-- --> | ||
| − | <span class='h3bb'>Sequence and Features</span> | + | <span class='h3bb'>Sequence and Features:</span> |
<partinfo>BBa_K1980010 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1980010 SequenceAndFeatures</partinfo> | ||
==Usage and Biology== | ==Usage and Biology== | ||
| + | Our project was to investigate a probiotic treatment for the copper-accumulation disorder: Wilson's disease. This required a system able to detect dietary copper, ideally in the range over which copper concentration changes after a meal (around 5-10μM). When copper is detected a copper chelating protein should be induced to lower the free copper concentration to prevent its absorption by the body. | ||
| + | ===CueR and pCopA=== | ||
<p><i>E. coli</i> cells use a protein called CueR to regulate the cytoplasmic copper concentration. CueR is a MerR-type regulator with an interesting mechanism of action whereby it can behave as a net activator or a net repressor under different copper concentrations through interaction with RNA polymerase<sup>(1)</sup>. CueR forms dimers consisting of three functional domains (a DNA-binding, a dimerisation and a metal-binding domain). The DNA binding domains bind to DNA inverted repeats called CueR boxes with the sequence:</p> | <p><i>E. coli</i> cells use a protein called CueR to regulate the cytoplasmic copper concentration. CueR is a MerR-type regulator with an interesting mechanism of action whereby it can behave as a net activator or a net repressor under different copper concentrations through interaction with RNA polymerase<sup>(1)</sup>. CueR forms dimers consisting of three functional domains (a DNA-binding, a dimerisation and a metal-binding domain). The DNA binding domains bind to DNA inverted repeats called CueR boxes with the sequence:</p> | ||
| Line 20: | Line 21: | ||
<p>This box is present at the promoter regions of the copper exporting ATPase CopA, some molybdenum cofactor synthesis genes and the periplasmic copper oxidase protein CueO.<sup>(2)</sup></p> | <p>This box is present at the promoter regions of the copper exporting ATPase CopA, some molybdenum cofactor synthesis genes and the periplasmic copper oxidase protein CueO.<sup>(2)</sup></p> | ||
| + | |||
| + | Here CueR is expressed from a constitutive promoter divergent from pCopA so that it is present in excess to that expressed from the <i>E. coli</i> genome. | ||
| + | |||
| + | ===Csp1=== | ||
| + | |||
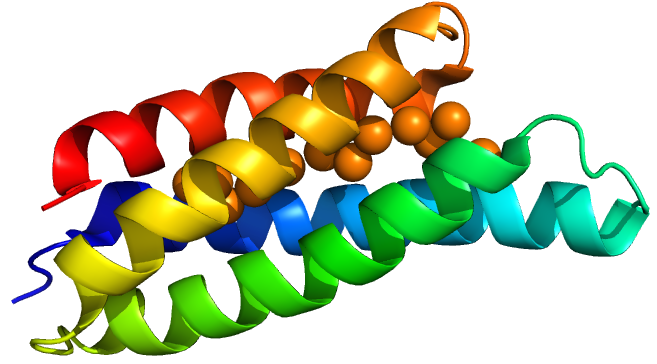
| + | <p>Copper storage protein 1 is a protein discovered in a methane-oxidizing alphaproteobacterium called <i>Methylosinus trichosporium OB3b. </i> (OB3b here stands for “oddball” strain 3b). This bacterium has a high demand for copper for use in its particular methane monoxygenase enzyme. Vita et al.<sup>(3)</sup> discovered Csp1 in 2015, characterised the protein’s copper affinity and obtained crystal structures with and without copper. | ||
| + | </p> | ||
| + | <p> | ||
| + | Csp1 is a tetramer of four-helix bundles. Each monomer can bind up to 13 Cu(I) ions meaning that the tetramer binds a maximum of 52 copper ions. Vita et al crystallised Csp1 with and without copper bound. The copper is bound inside the pre-folded helical bundles by Cys residues in contrast to metallothioneins, which are unstructured until they fold around metal ion clusters. Vita et al. found an average copper affinity of approximately 1x10<sup>17</sup>M<sup>-1</sup>. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | Csp1 has a signal peptide targeting it to the twin arginine translocation pathway (TAT). This means that it is likely a periplasmic protein. However they also found cytoplasmic homologues in many species challenging their and our assumption that only copper storage occurs in the periplasm due to copper toxicity. | ||
| + | </p> | ||
| + | <p> | ||
| + | We codon optimised Csp1 to <i>E. coli</i> and replaced the original TAT sequence with a TAT sequence from the <i>E. coli</i> protein CueO, which is also involved in copper regulation. | ||
| + | </p> | ||
| + | [[Image:T--Oxford--Sam-csp1-mono.jpg|400px|thumb|centre|'''Figure 1:''' A Csp1 monomer]] | ||
| + | |||
| + | ===Composite=== | ||
| + | |||
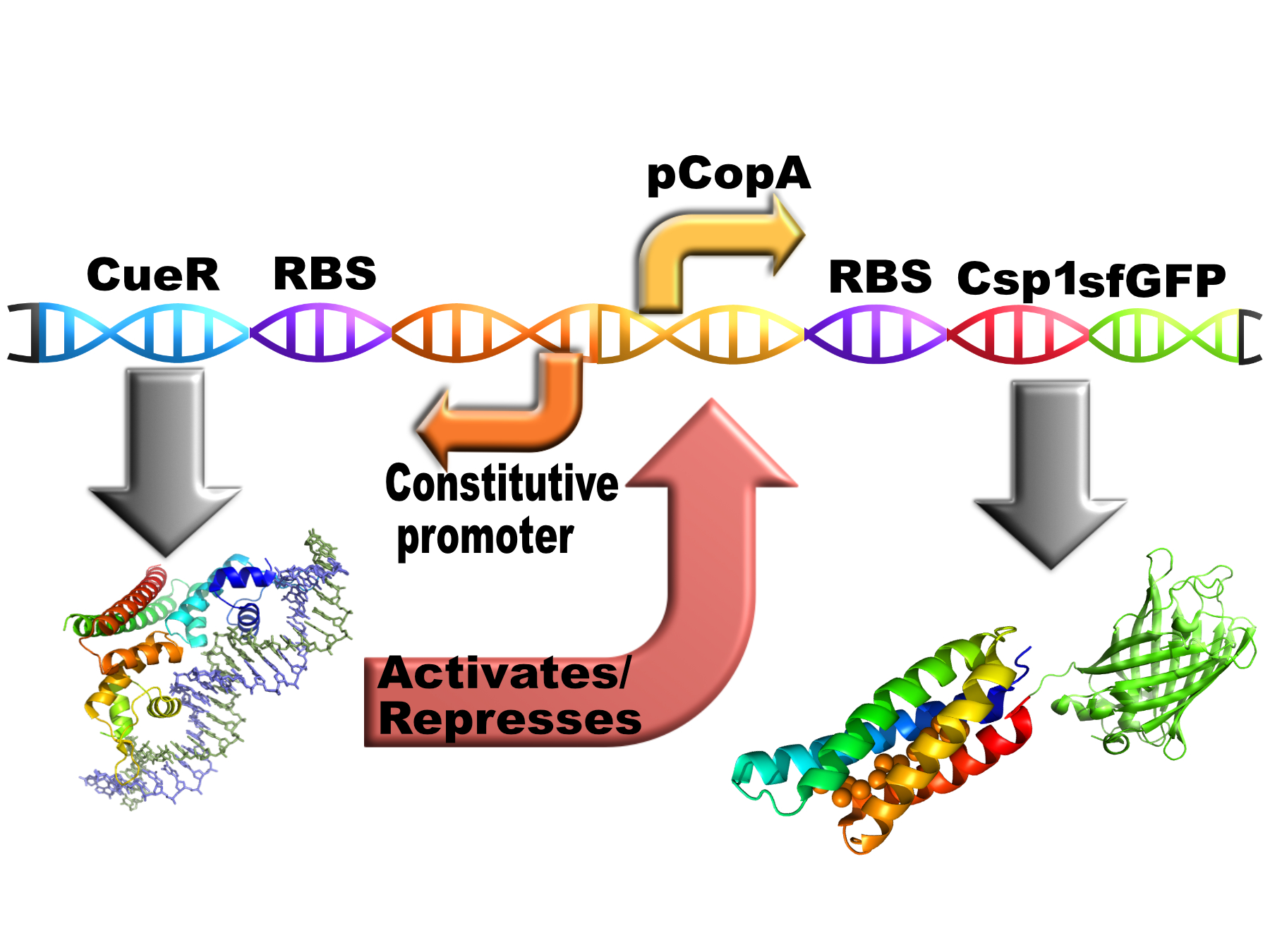
| + | <p> This is how the construct is intended to work <i>in vivo</i></p> | ||
| + | [[Image:T--Oxford--no10-4.jpeg|400px|thumb|center|Schematic of BBa K1980010]] | ||
| + | |||
| + | <p>Copper should induce the expression of the chelator via CueR. The chelator should reduce the free copper concentration therefore reducing it level of expression. This should in theory result in the free copper concentration falling to a low, stable level. </p> | ||
==Experience== | ==Experience== | ||
| − | <p>We cloned pCopA TAT Csp1 sfGFP with divergent expressed CueR from a gBlock into the shipping plasmid pSB1C3. <i>E. coli</i> strain MG1655 was transformed using the specific recombinant plasmid and a 5ml culture of a transformed colony was grown overnight | + | <p>We cloned pCopA TAT Csp1 sfGFP with divergent expressed CueR from a gBlock into the shipping plasmid pSB1C3. <i>E. coli</i> strain MG1655 was transformed using the specific recombinant plasmid and a 5ml culture of a transformed colony was grown overnight. The functionality of the construct was tested using plate reading, flow cytometry and microscopy</p> |
| − | |||
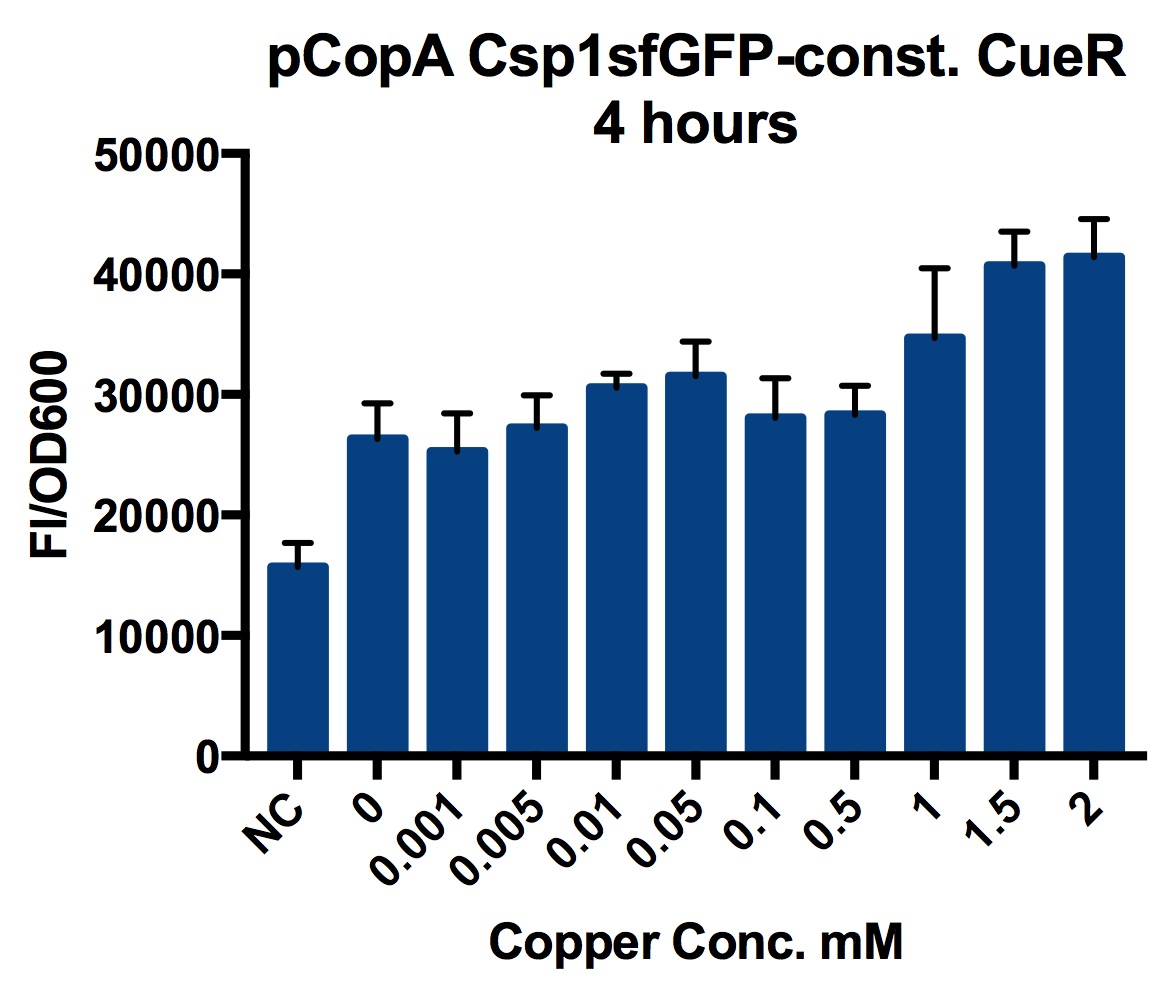
| − | [[Image:T--Oxford--no10- | + | ===Plate Reader=== |
| + | [[Image:T--Oxford--no10-1.jpeg|300px|thumb|right|The results of a plate reader experiment with different copper concentrations after four hours. Error bars show standard deviation of four repeats | ||
| − | + | ]] | |
| + | <p> | ||
| + | We tested the promoter with the fluorescent protein sfGFP (a form of GFP with excitation/emission maxima at 485-512nm and 520nm). | ||
| + | </p> | ||
| + | <p> | ||
| + | To account for the number of cells present at different copper concentrations and different times we measured the optical density (OD) as a proportional measure of the number of cells present. | ||
| + | </p> | ||
| + | <p> | ||
| + | A range of copper concentrations were prepared from stock solutions. A large volume plate was then prepared with 10μl of copper solution, 10μl of overnight culture and 980μl of broth with antibiotic. This resulted in a 1 in 100 dilution of the copper solutions prepared. | ||
| + | </p> | ||
| + | <p> | ||
| + | This large-volume plate was then centrifuged to mix the solutions and then 200μl transferred to a small-volume plate with a clear lid and then placed in the plate reader. </p> | ||
| + | <p> | ||
| + | Four biological repeats of this part at ten different copper concentrations were measured as well as four repeats of a negative control (The NC part from the Interlab study) and a positive control (The constitutive GFP, PC part from the Interlab study). | ||
| + | </p> | ||
| + | <p> | ||
| + | The plate reader measured the fluorescence and OD every ten minutes for at least 12 hours, shaking between measurements. | ||
| + | </p> | ||
| + | ===Flow cytometry=== | ||
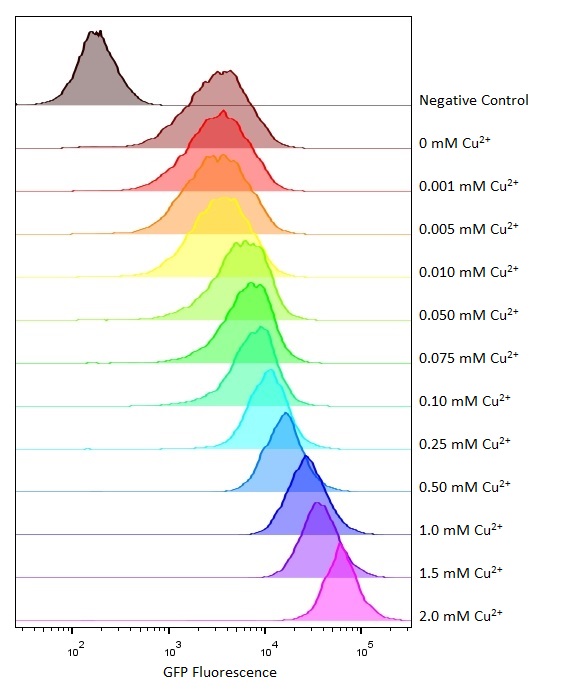
| + | [[Image:T--Oxford--no10-2.jpeg|300px|thumb|right|Flow cytometry measurements at the same copper concentration range]] | ||
| + | <p> | ||
| + | Flow cytometry is a technique whereby cells are passed individually into the path of a beam of light. The frequency range of incident light can be adjusted to allow excitation of specific fluorophores in the sample of cells. Downstream detectors can measure fluorescent emission, and this data can be used to quantify the amount of fluorophore in each cell.</p> | ||
| + | <p>To ensure comparability of experiments, all cells were grown for 3-4hrs (until entrance into the exponential growth phase) at 37°C and 225rpm shaking in the presence of the inducer before measurement. This allowed adequate time for activation of expression by the promotor systems. The negative control used in all cases were MG1655 bacteria containing an empty shipping plasmid. As these did not contain any fluorescent molecules, this population could be used to set the negative “gate” (i.e. the background fluorescence of the bacterial cells). Although the experiments were tedious as every sample had to be measured manually, the results were of remarkably high quality, were clearly interpretable, and fit very well with the other experimental data. | ||
| + | </p> | ||
| − | <p>Likely due to the expression problems of the Csp1 chelator in <i>E. coli</i>, it responded worse than the similar part with our MymTsfGFP fusion protein | + | |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===Microscopy=== | ||
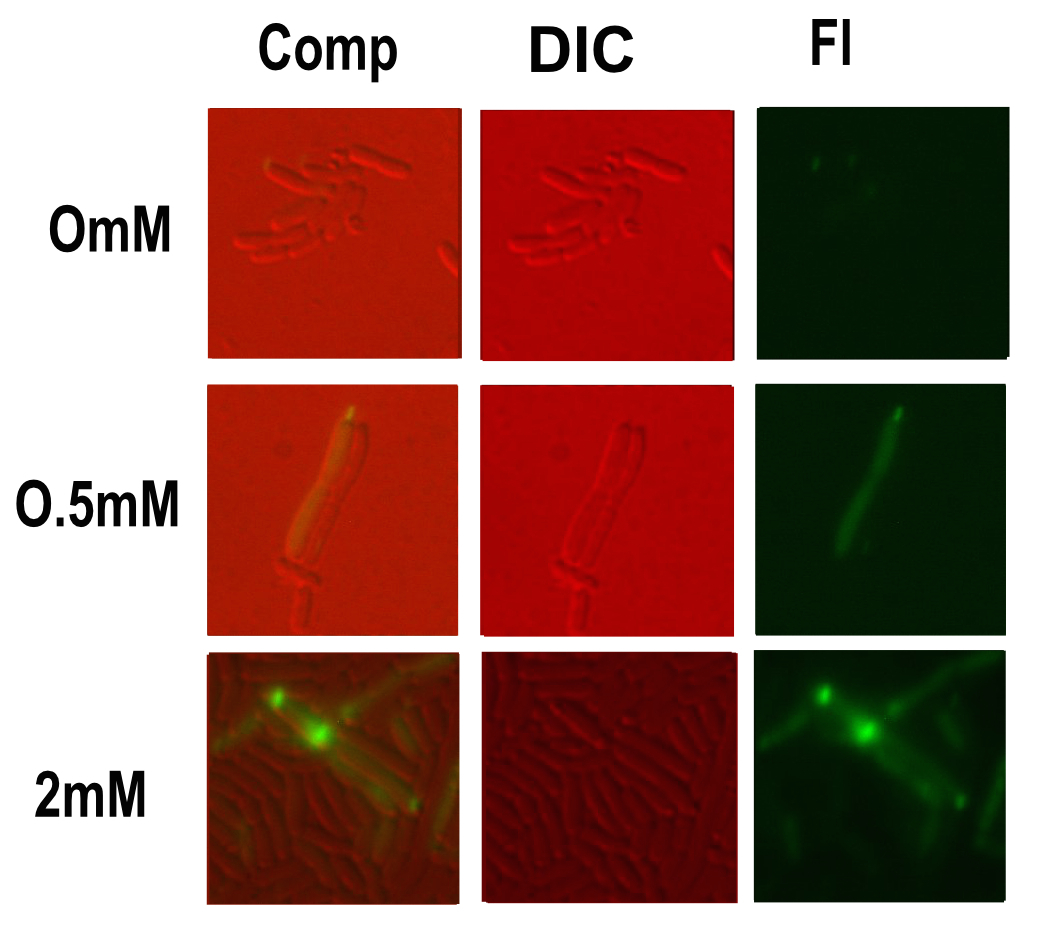
| + | [[Image:T--Oxford--no10-3.jpeg|300px|thumb|right|Microscopy performed on this part at different copper concentrations. Fluorescent, differential interference contrast channels and a composite shown]] | ||
| + | <p> | ||
| + | Microscopy was done in order to visually confirm the plate reader and flow cytometer experiments. | ||
| + | </p> | ||
| + | <p> | ||
| + | The experiment started with 5ml overnight cultures containing the appropriate antibiotic. Then in the morning 100μl of each colony was pipetted into 5ml of fresh LB with antibiotic and a range of copper concentrations and grown till the OD reached 0.4-0.6. | ||
| + | </p> | ||
| + | <p> | ||
| + | A flask of 1% agarose made with MilliQ was melted. 200μl of this was placed on a slide between two coverslips, flattened to get a nice smooth surface where the bacteria are immobile. 20μl of the culture are then added. The slide was then be viewed under a fluorescence microscope. | ||
| + | </p> | ||
| + | <p> | ||
| + | After finding the correct focal plane the slide was moved to find as many cells as possible to image. After focusing again an image of the DIC and fluorescence channels was obtained. | ||
| + | </p> | ||
| + | |||
| + | <p>Likely due to the expression problems of the Csp1 chelator in <i>E. coli</i>, it responded worse than the similar part with our MymTsfGFP fusion protein <html><a href="https://parts.igem.org/Part:BBa_K1980012">BBa_K1980012</a></html></p> | ||
| + | |||
| + | ===Absorbance Assay=== | ||
| + | |||
| + | <p>Separately we cloned Csp1sfGFP from Gblock (<html><a href="https://parts.igem.org/Part:BBa_K1980001">BBa_K1980001</a></html>) into the shipping vector then transferred it into the pBAD, arabinose-inducible commercial expression vector. This transfer necessitated the changing of the TAT sequence by the addition of a serine residue after the initiator methionine.</p> | ||
| + | |||
| + | <p>We were unable however to detect a drop in solution copper concentration due to copper chelation activity of Csp1sfGFP when expressed from pBAD in MG1655 <i>E. coli</i> strain, using an absorbance assay. Performing a similar procedure with this part also showed no measurable drop in copper concentration. | ||
| + | </p> | ||
| + | <p> | ||
| + | The assay used was the reagent Bathocuproine disulphonic Acid (BCS). BCS is colourless in the absence of Cu<sup>+</sup> but upon exposure to Cu<sup>+</sup>, BCS forms complexes with Cu<sup>+</sup> and absorbs strongly in the 480nm range. In our assays, at concentrations of 50µg/mL, the 480nm absorbance varied linearly with the Cu+ concentration from the detection limit of around 1µM Cu<sup>+</sup>, to approximately 20µM Cu<sup>+</sup>. As not only MymT and Csp1 bind copper in the Cu<sup>+</sup> form, but the assay also requires singly charged copper, the assays were optimised to include a suitable about of mild reducing agent to ensure reduction of the added CuSO<sub>4</sub> (releases Cu<sup>2+</sup>) to Cu<sup>+</sup>. After trying L(+)-Ascorbate and DL-Dithiothreitol, and L-Glutathione as candidates, L-Glutathione was selected as it was both mild enough to not damage biological material and efficient at reducing Cu<sup>2+</sup>. At >2-3 times the concentration of Cu<sup>2+</sup> in solution, L-Glutathione had maximum reductive activity against Cu<sup>2+</sup>.</p> | ||
| + | <p> Modelling by our team suggested that this was because insufficient protein could be expressed to chelate the amount needed to be detectable on the assay (1μM detection limit).</p> | ||
| + | <p>We purified this protein, from MG1655 cells expressing Csp1sfGFP from pBad, using Nickel affinity chromatography followed by dialysis. However were unable to detect copper chelation with the assay in our purified extracts when compared to a his-tagged GFP control. This is possibly because the flexible linked was susceptible to cleavage by proteases in the lysate meaning the protein we purified lacked the chelator portion. We attached the his tag to the C terminus of sfGFP rather than the N terminus of Csp1 in order to preserve the TAT sequence. </p> | ||
| + | |||
| + | ===Conclusions=== | ||
| + | <p>We were unable to demonstrate the ability of this system to buffer free copper concentration we believe that this is because the range of concentrations over which the system operates is below the detection limit of our absorbance assay. Future work should be done to optimise the system so it can operate over a wider range of copper concentrations. This could possibly be done by changing the promoter sequence or the RBS strengths.</p> | ||
==References== | ==References== | ||
| Line 39: | Line 131: | ||
<p>(1) Danya J. Martell, Chandra P. Joshi, Ahmed Gaballa, Ace George Santiago, Tai-Yen Chen, Won Jung, John D. Helmann, and Peng Chen (2015) “Metalloregulator CueR biases RNA polymerase’s kinetic sampling of dead-end or open complex to repress or activate transcription” Proc Natl Acad Sci U S A. 2015 Nov 3; 112(44): 13467–13472.</p> | <p>(1) Danya J. Martell, Chandra P. Joshi, Ahmed Gaballa, Ace George Santiago, Tai-Yen Chen, Won Jung, John D. Helmann, and Peng Chen (2015) “Metalloregulator CueR biases RNA polymerase’s kinetic sampling of dead-end or open complex to repress or activate transcription” Proc Natl Acad Sci U S A. 2015 Nov 3; 112(44): 13467–13472.</p> | ||
<p>(2) Yamamoto K, Ishihama A. (2005) “Transcriptional response of Escherichia coli to external copper.” Mol Microbiol. 2005 Apr;56(1):215-27.</p> | <p>(2) Yamamoto K, Ishihama A. (2005) “Transcriptional response of Escherichia coli to external copper.” Mol Microbiol. 2005 Apr;56(1):215-27.</p> | ||
| + | <p>(3) Nicolas Vita, Semeli Platsaki, Arnaud Basle, Stephen J. Allen, Neil G. Paterson, Andrew T. Crombie, J. Colin Murrell, Kevin J.Waldron & Christopher Dennison (2015) | ||
| + | “A four-helix bundle stores copper for methane oxidation”, Nature 525 issue 7567 pg. 140-143 doi:10.1038/nature14854</p> | ||
| + | |||
| + | |||
| + | <p style="text-align: right"><i>Author: Andreas Hadjicharalambous and Sam Garforth</i></p> | ||
Latest revision as of 20:59, 24 October 2016
pCopA TAT Csp1 sfGFP with divergent CueR
Description
This part contains the bacterial copper chelator Csp1 (Copper Storage Protein 1) with a C-terminal sfGFP tag (connected with short hydrophilic flexible linker) behind the pCopA promoter with divergently expressed CueR (a bacterial copper sensitive repressor/activator).
Sequence and Features:
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 438
Illegal NheI site found at 461 - 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 1466
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1028
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Our project was to investigate a probiotic treatment for the copper-accumulation disorder: Wilson's disease. This required a system able to detect dietary copper, ideally in the range over which copper concentration changes after a meal (around 5-10μM). When copper is detected a copper chelating protein should be induced to lower the free copper concentration to prevent its absorption by the body.
CueR and pCopA
E. coli cells use a protein called CueR to regulate the cytoplasmic copper concentration. CueR is a MerR-type regulator with an interesting mechanism of action whereby it can behave as a net activator or a net repressor under different copper concentrations through interaction with RNA polymerase(1). CueR forms dimers consisting of three functional domains (a DNA-binding, a dimerisation and a metal-binding domain). The DNA binding domains bind to DNA inverted repeats called CueR boxes with the sequence:
CCTTCCNNNNNNGGAAGG
This box is present at the promoter regions of the copper exporting ATPase CopA, some molybdenum cofactor synthesis genes and the periplasmic copper oxidase protein CueO.(2)
Here CueR is expressed from a constitutive promoter divergent from pCopA so that it is present in excess to that expressed from the E. coli genome.
Csp1
Copper storage protein 1 is a protein discovered in a methane-oxidizing alphaproteobacterium called Methylosinus trichosporium OB3b. (OB3b here stands for “oddball” strain 3b). This bacterium has a high demand for copper for use in its particular methane monoxygenase enzyme. Vita et al.(3) discovered Csp1 in 2015, characterised the protein’s copper affinity and obtained crystal structures with and without copper.
Csp1 is a tetramer of four-helix bundles. Each monomer can bind up to 13 Cu(I) ions meaning that the tetramer binds a maximum of 52 copper ions. Vita et al crystallised Csp1 with and without copper bound. The copper is bound inside the pre-folded helical bundles by Cys residues in contrast to metallothioneins, which are unstructured until they fold around metal ion clusters. Vita et al. found an average copper affinity of approximately 1x1017M-1.
Csp1 has a signal peptide targeting it to the twin arginine translocation pathway (TAT). This means that it is likely a periplasmic protein. However they also found cytoplasmic homologues in many species challenging their and our assumption that only copper storage occurs in the periplasm due to copper toxicity.
We codon optimised Csp1 to E. coli and replaced the original TAT sequence with a TAT sequence from the E. coli protein CueO, which is also involved in copper regulation.
Composite
This is how the construct is intended to work in vivo
Copper should induce the expression of the chelator via CueR. The chelator should reduce the free copper concentration therefore reducing it level of expression. This should in theory result in the free copper concentration falling to a low, stable level.
Experience
We cloned pCopA TAT Csp1 sfGFP with divergent expressed CueR from a gBlock into the shipping plasmid pSB1C3. E. coli strain MG1655 was transformed using the specific recombinant plasmid and a 5ml culture of a transformed colony was grown overnight. The functionality of the construct was tested using plate reading, flow cytometry and microscopy
Plate Reader
We tested the promoter with the fluorescent protein sfGFP (a form of GFP with excitation/emission maxima at 485-512nm and 520nm).
To account for the number of cells present at different copper concentrations and different times we measured the optical density (OD) as a proportional measure of the number of cells present.
A range of copper concentrations were prepared from stock solutions. A large volume plate was then prepared with 10μl of copper solution, 10μl of overnight culture and 980μl of broth with antibiotic. This resulted in a 1 in 100 dilution of the copper solutions prepared.
This large-volume plate was then centrifuged to mix the solutions and then 200μl transferred to a small-volume plate with a clear lid and then placed in the plate reader.
Four biological repeats of this part at ten different copper concentrations were measured as well as four repeats of a negative control (The NC part from the Interlab study) and a positive control (The constitutive GFP, PC part from the Interlab study).
The plate reader measured the fluorescence and OD every ten minutes for at least 12 hours, shaking between measurements.
Flow cytometry
Flow cytometry is a technique whereby cells are passed individually into the path of a beam of light. The frequency range of incident light can be adjusted to allow excitation of specific fluorophores in the sample of cells. Downstream detectors can measure fluorescent emission, and this data can be used to quantify the amount of fluorophore in each cell.
To ensure comparability of experiments, all cells were grown for 3-4hrs (until entrance into the exponential growth phase) at 37°C and 225rpm shaking in the presence of the inducer before measurement. This allowed adequate time for activation of expression by the promotor systems. The negative control used in all cases were MG1655 bacteria containing an empty shipping plasmid. As these did not contain any fluorescent molecules, this population could be used to set the negative “gate” (i.e. the background fluorescence of the bacterial cells). Although the experiments were tedious as every sample had to be measured manually, the results were of remarkably high quality, were clearly interpretable, and fit very well with the other experimental data.
Microscopy
Microscopy was done in order to visually confirm the plate reader and flow cytometer experiments.
The experiment started with 5ml overnight cultures containing the appropriate antibiotic. Then in the morning 100μl of each colony was pipetted into 5ml of fresh LB with antibiotic and a range of copper concentrations and grown till the OD reached 0.4-0.6.
A flask of 1% agarose made with MilliQ was melted. 200μl of this was placed on a slide between two coverslips, flattened to get a nice smooth surface where the bacteria are immobile. 20μl of the culture are then added. The slide was then be viewed under a fluorescence microscope.
After finding the correct focal plane the slide was moved to find as many cells as possible to image. After focusing again an image of the DIC and fluorescence channels was obtained.
Likely due to the expression problems of the Csp1 chelator in E. coli, it responded worse than the similar part with our MymTsfGFP fusion protein BBa_K1980012
Absorbance Assay
Separately we cloned Csp1sfGFP from Gblock (BBa_K1980001) into the shipping vector then transferred it into the pBAD, arabinose-inducible commercial expression vector. This transfer necessitated the changing of the TAT sequence by the addition of a serine residue after the initiator methionine.
We were unable however to detect a drop in solution copper concentration due to copper chelation activity of Csp1sfGFP when expressed from pBAD in MG1655 E. coli strain, using an absorbance assay. Performing a similar procedure with this part also showed no measurable drop in copper concentration.
The assay used was the reagent Bathocuproine disulphonic Acid (BCS). BCS is colourless in the absence of Cu+ but upon exposure to Cu+, BCS forms complexes with Cu+ and absorbs strongly in the 480nm range. In our assays, at concentrations of 50µg/mL, the 480nm absorbance varied linearly with the Cu+ concentration from the detection limit of around 1µM Cu+, to approximately 20µM Cu+. As not only MymT and Csp1 bind copper in the Cu+ form, but the assay also requires singly charged copper, the assays were optimised to include a suitable about of mild reducing agent to ensure reduction of the added CuSO4 (releases Cu2+) to Cu+. After trying L(+)-Ascorbate and DL-Dithiothreitol, and L-Glutathione as candidates, L-Glutathione was selected as it was both mild enough to not damage biological material and efficient at reducing Cu2+. At >2-3 times the concentration of Cu2+ in solution, L-Glutathione had maximum reductive activity against Cu2+.
Modelling by our team suggested that this was because insufficient protein could be expressed to chelate the amount needed to be detectable on the assay (1μM detection limit).
We purified this protein, from MG1655 cells expressing Csp1sfGFP from pBad, using Nickel affinity chromatography followed by dialysis. However were unable to detect copper chelation with the assay in our purified extracts when compared to a his-tagged GFP control. This is possibly because the flexible linked was susceptible to cleavage by proteases in the lysate meaning the protein we purified lacked the chelator portion. We attached the his tag to the C terminus of sfGFP rather than the N terminus of Csp1 in order to preserve the TAT sequence.
Conclusions
We were unable to demonstrate the ability of this system to buffer free copper concentration we believe that this is because the range of concentrations over which the system operates is below the detection limit of our absorbance assay. Future work should be done to optimise the system so it can operate over a wider range of copper concentrations. This could possibly be done by changing the promoter sequence or the RBS strengths.
References
(1) Danya J. Martell, Chandra P. Joshi, Ahmed Gaballa, Ace George Santiago, Tai-Yen Chen, Won Jung, John D. Helmann, and Peng Chen (2015) “Metalloregulator CueR biases RNA polymerase’s kinetic sampling of dead-end or open complex to repress or activate transcription” Proc Natl Acad Sci U S A. 2015 Nov 3; 112(44): 13467–13472.
(2) Yamamoto K, Ishihama A. (2005) “Transcriptional response of Escherichia coli to external copper.” Mol Microbiol. 2005 Apr;56(1):215-27.
(3) Nicolas Vita, Semeli Platsaki, Arnaud Basle, Stephen J. Allen, Neil G. Paterson, Andrew T. Crombie, J. Colin Murrell, Kevin J.Waldron & Christopher Dennison (2015) “A four-helix bundle stores copper for methane oxidation”, Nature 525 issue 7567 pg. 140-143 doi:10.1038/nature14854
Author: Andreas Hadjicharalambous and Sam Garforth