Difference between revisions of "Part:BBa I0500:Experience"
(→Characterization by BNU-China 2022) |
|||
| (64 intermediate revisions by 9 users not shown) | |||
| Line 170: | Line 170: | ||
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
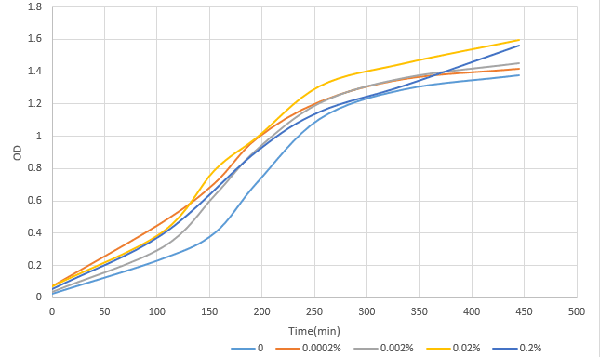
| − | + | [http://2016.igem.org/Team:OUC-China OUC-China] characterized BBa_I0500 of different concentrations of L-arabinose on the transcriptional level. We found that the strength of I0500 was mainly measured on the translational level before. As far as we are concerned, it is the transcriptional level that can directly reflect I0500’s original strength without interfering by context. In order to verify it more accurately, we tested it on the transcriptional level by extracting RNA and measured it by qPCR. We constructed I0500 with GFP(E0040) and transformed into TOP10F’. Firstly , we plated on LB agar with chloramphenicol and individual colonies were subsequently grown overnight in LB-medium. Cultures were back diluted 1:100 into 100mL fresh LB-medium with chloramphenicol, adding 0,0.0002%,0.002%,0.02%,0.2% (m/V)L-arabinose. After growing for 18 hours, we extracted RNA and measured it. At the same time, we measured the fluorescence of GFP(485nm/520nm). Besides, we also measured OD over time to check whether L-arabinose promotes bacteria’s growth.<br> | |
| − | + | [[File:T--OUC-China--parts-333.png|center|thumb|400px|Figure1 OD after induction with different concentration of L-arabinose over time]] | |
| − | [[File:T--OUC-China--parts-333.png|center|thumb| | + | [[File:T--OUC-China--parts-222.png|center|thumb|400px|Figure2 The relative expression level of GFP after induction with different concentration of L-arabinose.]] |
| − | [[File:T--OUC-China--parts-222.png|center|thumb| | + | 1. 100mlLB,blank<br> |
| − | + | 2. 100mlLB,0.0002%arobinose<br> | |
| − | 1. 100mlLB,blank | + | 3. 100mlLB,0.002%arobinose<br> |
| − | 2. 100mlLB,0.0002%arobinose | + | 4. 100mlLB,0.02%arobinose<br> |
| − | 3. 100mlLB,0.002%arobinose | + | 5. 100mlLB,0.2%arobinose<br> |
| − | 4. 100mlLB,0.02%arobinose | + | |
| − | 5. 100mlLB,0.2%arobinose | + | |
According to our data analysis, when induced with 0.02% arabinose, our strain TOP10 was effected better than that induced with 0.2% according to the previous research. We infer that 0.2% arabinose may affect the growth of our strain. So, in the future, we suggest others to use this concentration to induce the TOP10. | According to our data analysis, when induced with 0.02% arabinose, our strain TOP10 was effected better than that induced with 0.2% according to the previous research. We infer that 0.2% arabinose may affect the growth of our strain. So, in the future, we suggest others to use this concentration to induce the TOP10. | ||
|} | |} | ||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_I0500 AddReview 4</partinfo> | ||
| + | <I>[http://2017.igem.org/Team:Glasgow]Team Glasgow 2017</I> | ||
| + | |width='60%' valign='top'| | ||
| + | |||
| + | [http://2017.igem.org/Team:Glasgow Team Glasgow 2017] improved this part by splitting its constituent parts into two separate BioBricks: [https://parts.igem.org/Part:BBa_K2442101 BBa_K2442101] encoding minimal pBAD and [https://parts.igem.org/Part:BBa_K2442104 BBa_K2442104] encoding AraC under regulation of LacI-regulated promoter. This allows for greater control over arabinose-inducible systems. Placing AraC under control of LacI regulated promoter (R0011) allows to induce expression of AraC only when required, taking translational load off grown cells. R0011 is inducible by IPTG. AraC coding sequence alone is also available as part [https://parts.igem.org/Part:BBa_K2442103 BBa_K2442103], which can be placed under other promoter of choice. | ||
| + | <p>We isolated the minimal DNA sequence from the native pBAD that is sufficient to retain its function. The promoter retains all the operator sites O<sub>1</sub>, O<sub>2</sub>, I<sub>1</sub> and I<sub>2</sub> required for binding of AraC. As some of these sites lie within the PC promoter and overlap with AraC coding sequence, the minimal pBAD has the ''araC'' start codon ATG mutated to AGT to prevent the protein from being made. See part [https://parts.igem.org/Part:BBa_K2442101 BBa_K2442101] and our [http://2017.igem.org/Team:Glasgow/araC wiki] for details. </p> | ||
| + | <p> We tested pBAD with the use of pBAD+GFP reporter plasmid ([https://parts.igem.org/Part:BBa_K2442102 BBa_K2442102]) by measuring fluorescence. Activity of the promoter was tested in presence of arabinose, xylose, decanal and no inducer (Fig. 1). When transformed into AraC-positive ''E. coli'' strains such as DH5α, pBAD is sufficiently activated with just the chromosomal copy of ''araC'' and does not require introduction of ''araC'' from external plasmid. Moreover, DH5α cells carrying K2442102 showed fluorescence in presence of arabinose, but no significant increase in fluorescence in other conditions, showing high specificity of AraC and minimal leakage of minimal pBAD. DH5α carrying pBAD+GFP reporter on a high copy number plasmid (pSB1C3) [https://parts.igem.org/Part:BBa_K2442102 BBa_K2442102] showed 2000x fold increase in fluorescence on arabinose plate compared to empty cells (Fig. 1). This proves significant improvement of part [https://parts.igem.org/Part:BBa_I0500 BBa_I0500]. DH5α cells carrying minimal pBAD and I13500 on a low copy number plasmid pSB3K3 instead of pSB1C3 also showed expression of GFP only under arabinose conditions. However, levels of fluorescence were much lower, suggesting that the level of GFP production is dependent on the plasmid copy number and must be taken into account in further experiments. In AraC-negative strains, AraC expressed from [https://parts.igem.org/Part:BBa_K2442104 BBa_K2442104] induces pBAD with similar efficiency. </p> <p> Overall, results show that both AraC and arabinose are necessary to activate pBAD. Efficiency of minimal pBAD is demonstrated by 2000x fold increase in fluorescence of cells carrying the reporter plasmid. | ||
| + | |||
| + | [[Image:T-Glasgow-I0500-Graph.png|centre|thumb|800px|'''Figure 1:Average relative fluorescence over optical density at hour 8. Shows fluorescence levels under: no additive, 40mM Arabinose, 40mM Xylose and 2mM decanal. The ''E. coli'' strains which the plasmids have been transformed into are DS941 or DH5α (specified). DH5α is AraC-positive. DS941 is AraC-negative. BioBricks (K244210..) specified. All parts are in plasmid backbone pSB1C3 unless specified otherwise (pSB3k3). K2442102: reporter plasmid pBADmin+GFP. K2442104: regulatory plasmid R0011+B0032+AraC''']] | ||
| + | |||
| + | |||
| + | |}; | ||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_I0500 AddReview 5</partinfo> | ||
| + | <I>BHSF_ND iGEM 2019</I> | ||
| + | |width='60%' valign='top'| | ||
| + | We mainly focused on two types of inducible transcriptional promoter—Xyls and pBAD. | ||
| + | The Xyls protein is the positive transcription regulator of the TOL plasmid meta-cleavage pathway operon Pm. Xyls belongs to the AraC family of transcriptional regulators and exhibits an N-terminal domain involved in effector recognition, and a C-terminal domain.The activator Xyls is usually is usually activated by the benzoate and its derivatives (3MBz in our design). | ||
| + | The pBAD promoter holds great potential to control the expression of genes that require tight regulation, as it is capable of both an induction and repression. The pBAD promoter exhibits an almost all-or-none behavior upon induction with arabinose when located on a high copy vector, but allows for gradual induction when cloned into a low copy vector. Based on these research findings, a low copy vector was used to investigate the ability to inhibit gene expression subsequent to induction of pBAD. | ||
| + | |||
| + | As the result shows, both system exhibit relative high level of fluorescence compared to our backbone design( BN006/Contains BBa_K3202029) which suggest the two inducible transcription system function properly. | ||
| + | |||
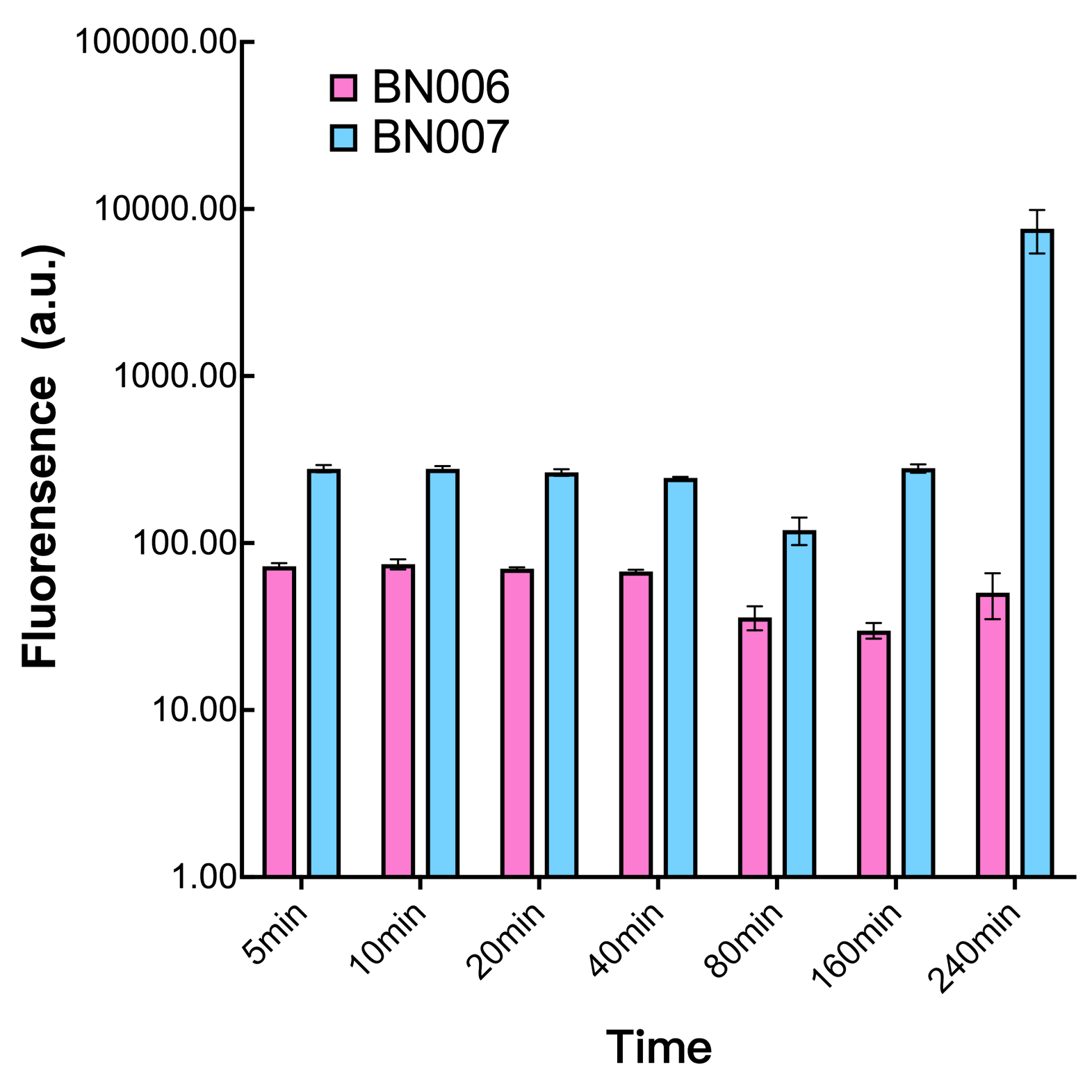
| + | [[File:T--BHSF_ND--Leakage experiment 1.png|429px|thumb|left|alt=Comparison between BN006/Contains BBa_K3202029 and BN007/Contains BBa_K3202030|Figure 1: pBAD(BN007/Contains BBa_K3202030) induction curve at different concentrations after 4 hours compared to backbone.(BN006/Contains BBa_K3202029)]] | ||
| + | |||
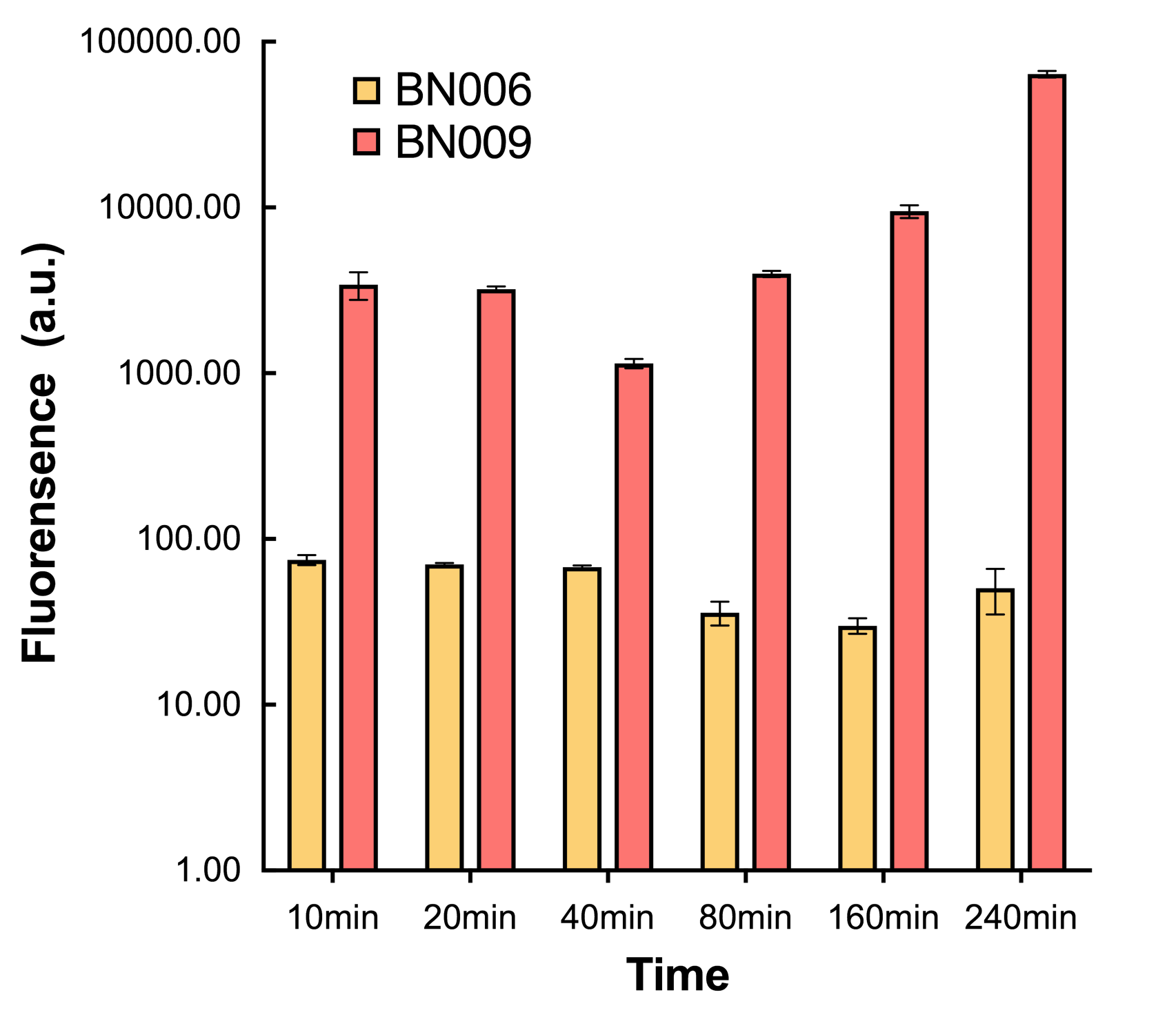
| + | [[File:T--BHSF_ND--Leakage experiment 2.png|435px|thumb|right|alt=Comparison between BN006/Contains BBa_K3202029 and BN009/Contains BBa_K3202031|Figure 2: Xyls(BN009/Contains BBa_K3202031) induction curve at different concentrations after 4 hours compared to backbone(BN006/Contains BBa_K3202029)]] | ||
| + | |||
| + | [[File:T--BHSF_ND--Leakage experiment 3.png|429px|thumb|left|alt=Comparison between BN006/Contains BBa_K3202029 and BN007/Contains BBa_K3202030|Figure3: Time induction curve pBAD(BN007/Contains BBa_K3202030) at the concentration of 1mM compared to backbone (BN006/Contains BBa_K3202029)]] | ||
| + | |||
| + | [[File:T--BHSF_ND--Leakage experiment 4.png|435px|thumb|right|alt=Comparison between BN006/Contains BBa_K3202029 and BN009/Contains BBa_K3202031|Figure4: Time induction curve for Xyls(BN009/Contains BBa_K3202031) at the concentration of 1mM compared to backbone (BN006/Contains BBa_K3202029)]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | === Results === | ||
| + | |||
| + | In our experiment of leakage testing, we used low-copying plasmid backbone PSB4K5, with a PSC101 origin of replication. | ||
| + | |||
| + | Two inducers with their respective promoters (AraC & pBAD; Xyls & Pm) are coupled with sfGFP to see if there is actually expression leakage when inducer is present quantitatively through flow cytometry. We tested fluorescence under different inducer concentration at the given time of 240 minutes and under different time stage at a given inducer concentration. Theoretically when inducers (AraC and 3MBz ) are present, promoters (pBAD and Pm) are initiated therefore both sfGFP are expressed; while sfGFP shouldn’t be expressed if inducer is absent. | ||
| + | When measured at the end of 240 minutes without inducer, BN009/Contains BBa_K3202031(Xyls+sfGFP) has a basal leakage of 968 a.u. compared to BN006/Contains BBa_K3202029(backbone without GFP). BN007/Contains BBa_K3202030(pBAD+sfGFP) also showed a leakage of 90 a.u. The result suggested that both system have noticeable leakage and Xyls has much higher leakage. | ||
| + | |||
| + | When we gradually increased inducer concentration to 1mM, the fluorescence of BN006/Contains BBa_K3202029 stays at a constant level, whereas the fluorescence of both Xyls and pBAD have increased exponentially. This is another sign that both system function properly. | ||
| + | |||
| + | We found an intriguing result when measuring the fluorescence of the two system under a constant inducer concentration of 1mM. During the first 160 minutes, the fluorescence of BN007/Contains BBa_K3202030 is 200 a.u. higher than BN006/Contains BBa_K3202029. However, at 240 minutes, the fluorescence value suddenly increased to 7641 a.u., which is 7590 a.u., higher than the backbone. Same thing happened to BN009/Contains BBa_K3202031. Therefore, in the subsequent experiments, systems are induced to up to 4 hours to ensure the accuracy of the experiment. Results shown above verified the presence of expression leakage of both systems when inducer is not expressed through the detectable fluorescence of reporter protein using flow cytometry. | ||
| + | |} | ||
| + | |||
| + | = Contribution iGEM TU-Eindhoven 2017 [http://2017.igem.org/Team:TU-Eindhoven] = | ||
| + | == Method == | ||
| + | We first tested three different temperatures, being 20, 25 and 37 degree Celsius. From our fist basic measurements, we discovered that expression was more efficient at a temperature of 37 degree Celsius and continued with this setting on our incubator. | ||
| + | The next step was consequent adding arabinose to the cultures, as E. coli BL21(DE3) is a strain that can break down arabinose, and for an optimal expression, it is better to have a high enough constant concentration. | ||
| + | We tested an addition of every 60 minutes and every 90 minutes, with varying concentrations of arabinose. | ||
| + | The percentages that are depicted in the figures are the total concentration of arabinose in the whole culture. The indication half means that we added 50% of the original added amount of arabinose compared with the first addition. This was done because we expected that a higher initial dose was needed to induce the expression, but that not all added arabinose would be broken down, and we therefore didn't need to add a high amount of arabinose. | ||
| + | |||
| + | == pBAD expression of fluorophore mCherry == | ||
| + | |||
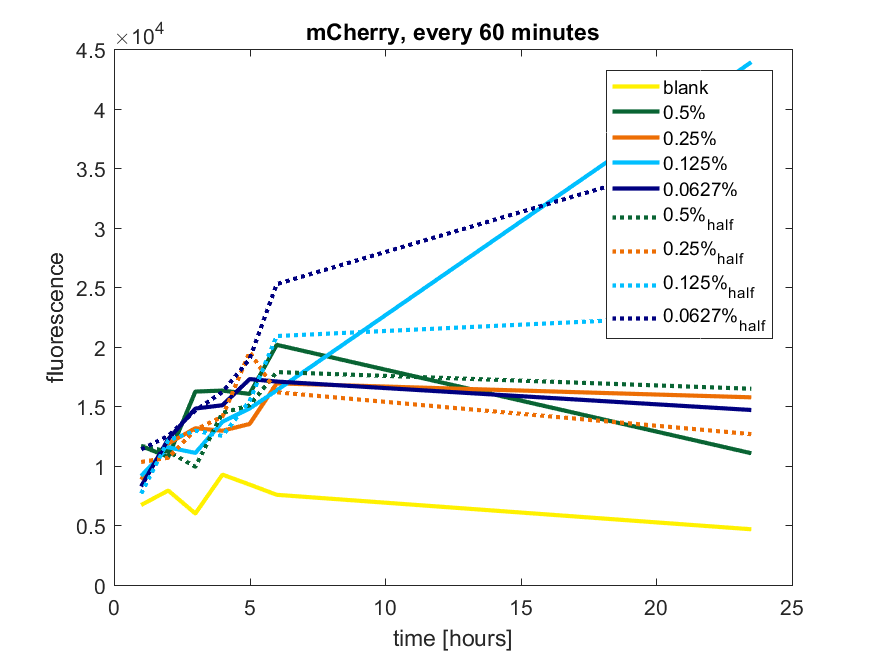
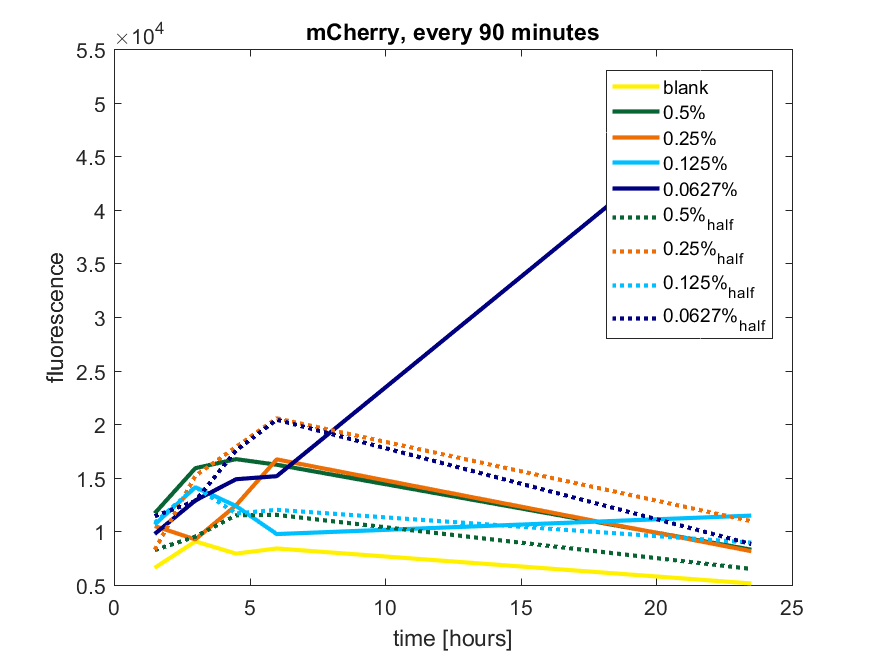
| + | The results of all our tries are depicted in Figure a1 & a2 with the best results in a3. | ||
| + | |||
| + | [[File:T-TU-Eindhoven--pBAD_vector_mCherry_60min.png]] | ||
| + | |||
| + | ''Figure a1: pBAD expression of mCherry in BL21(DE3) with every 60 minutes an addition of arabinose'' | ||
| + | |||
| + | [[File:T-TU-Eindhoven--pBAD_vector_mCherry_90min.png]] | ||
| + | |||
| + | ''Figure a2: pBAD expression of mCherry in BL21(DE3) with every 90 minutes an addition of arabinose'' | ||
| + | |||
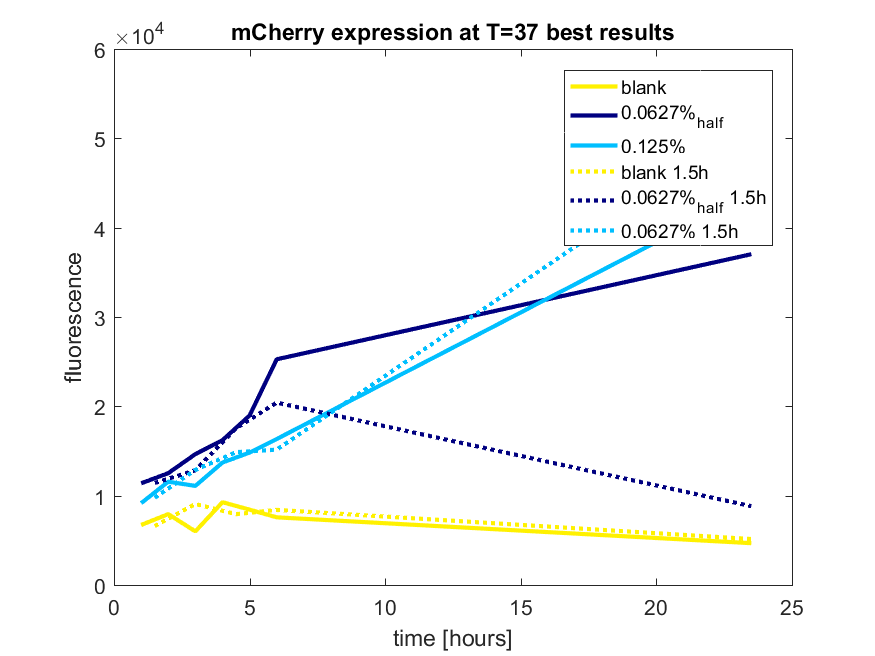
| + | [[File:T-TU-Eindhoven--pBAD_vector_mCherry_best_results.png]] | ||
| + | |||
| + | ''Figure a3: pBAD expression of mCHerry in BL21(DE3) most successful addition methods'' | ||
| + | |||
| + | == pBAD expression of fluorophore GFP == | ||
| + | |||
| + | The results of all our tries are depicted in Figure b1 & b2 with the best results in b3. | ||
| + | |||
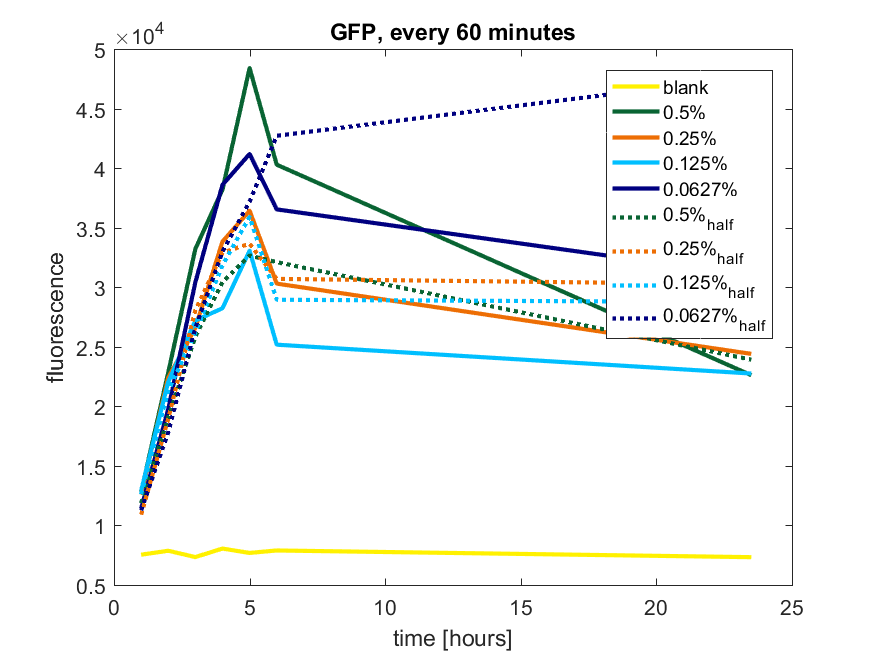
| + | [[File:T-TU-Eindhoven--pBAD_vector_GFP_60min.png]] | ||
| + | |||
| + | ''Figure b1: pBAD expression of GFP in BL21(DE3) with every 60 minutes an addition of arabinose'' | ||
| + | |||
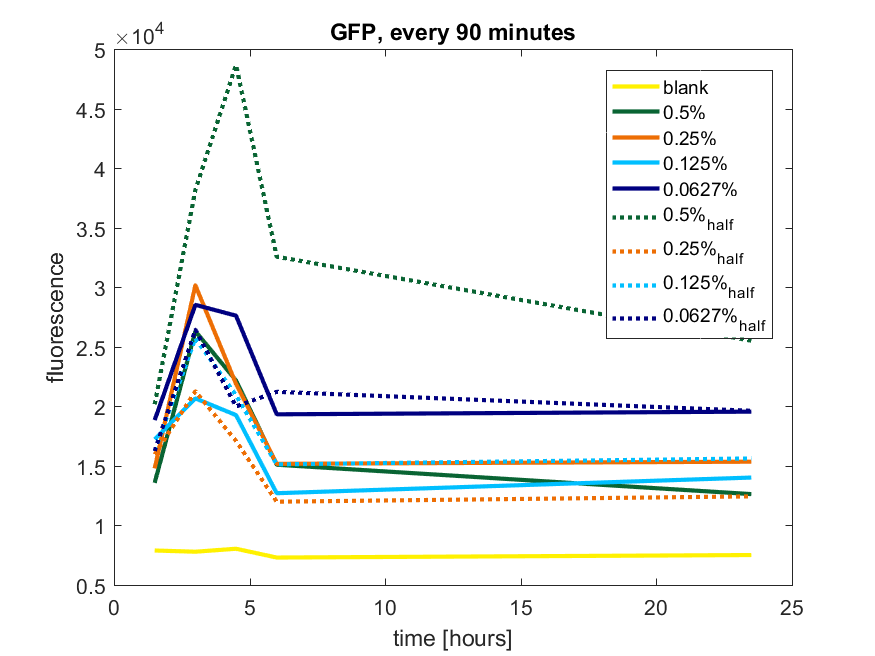
| + | [[File:T-TU-Eindhoven--pBAD_vector_GFP_90min.png]] | ||
| + | |||
| + | ''Figure b2: pBAD expression of GFP in BL21(DE3) with every 90 minutes an addition of arabinose'' | ||
| + | |||
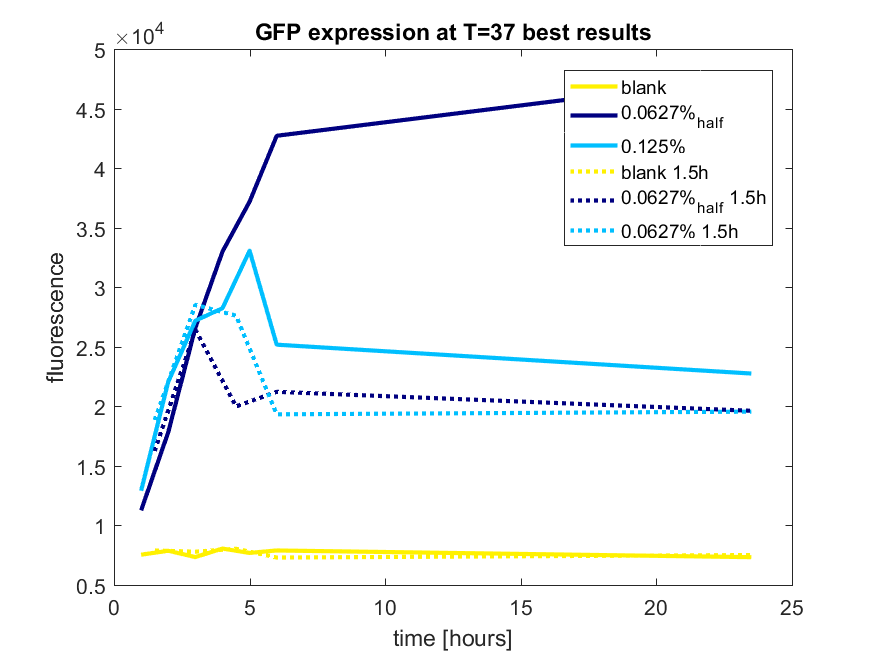
| + | [[File:T-TU-Eindhoven--pBAD_vector_GFP_best_results.png]] | ||
| + | |||
| + | ''Figure b3: pBAD expression of GFP in BL21(DE3) most successful addition methods'' | ||
| + | |||
| + | == pBAD expression conclusion == | ||
| + | |||
| + | The best results are depicted in Figure c. | ||
| + | |||
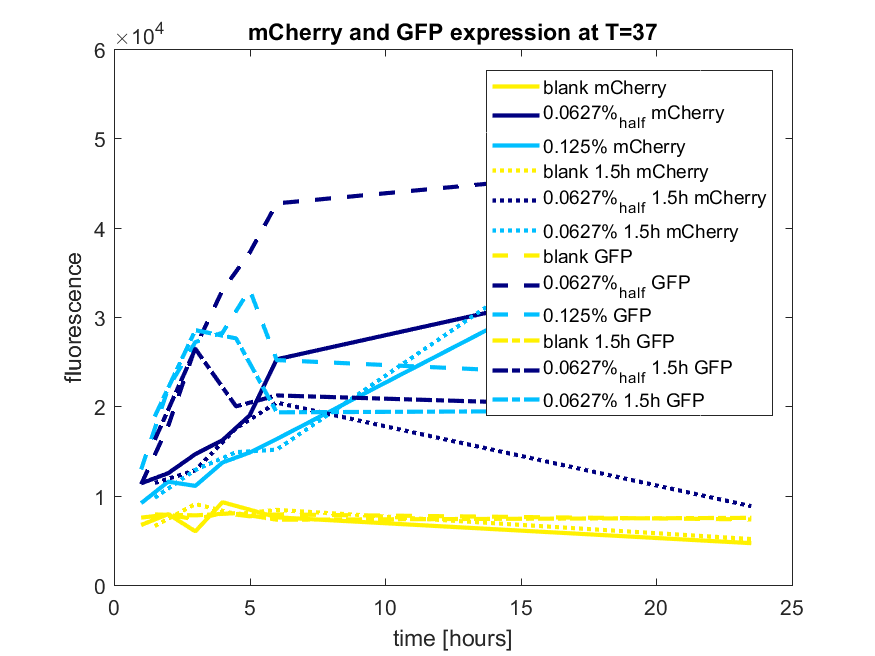
| + | [[File:T-TU-Eindhoven--pBAD_vector_mCherry_and_GFP_best_results.png]] | ||
| + | |||
| + | ''Figure c: pBAD expression of mCherry and GFP in BL21(DE3) most successful addition methods'' | ||
| + | |||
| + | <h2><span class="mw-headline" id="2021 Team Leiden">2021 Team Leiden</span><span class="mw-editsection"></span></h2> | ||
| + | |||
| + | https://2021.igem.org/Team:Leiden | ||
| + | |||
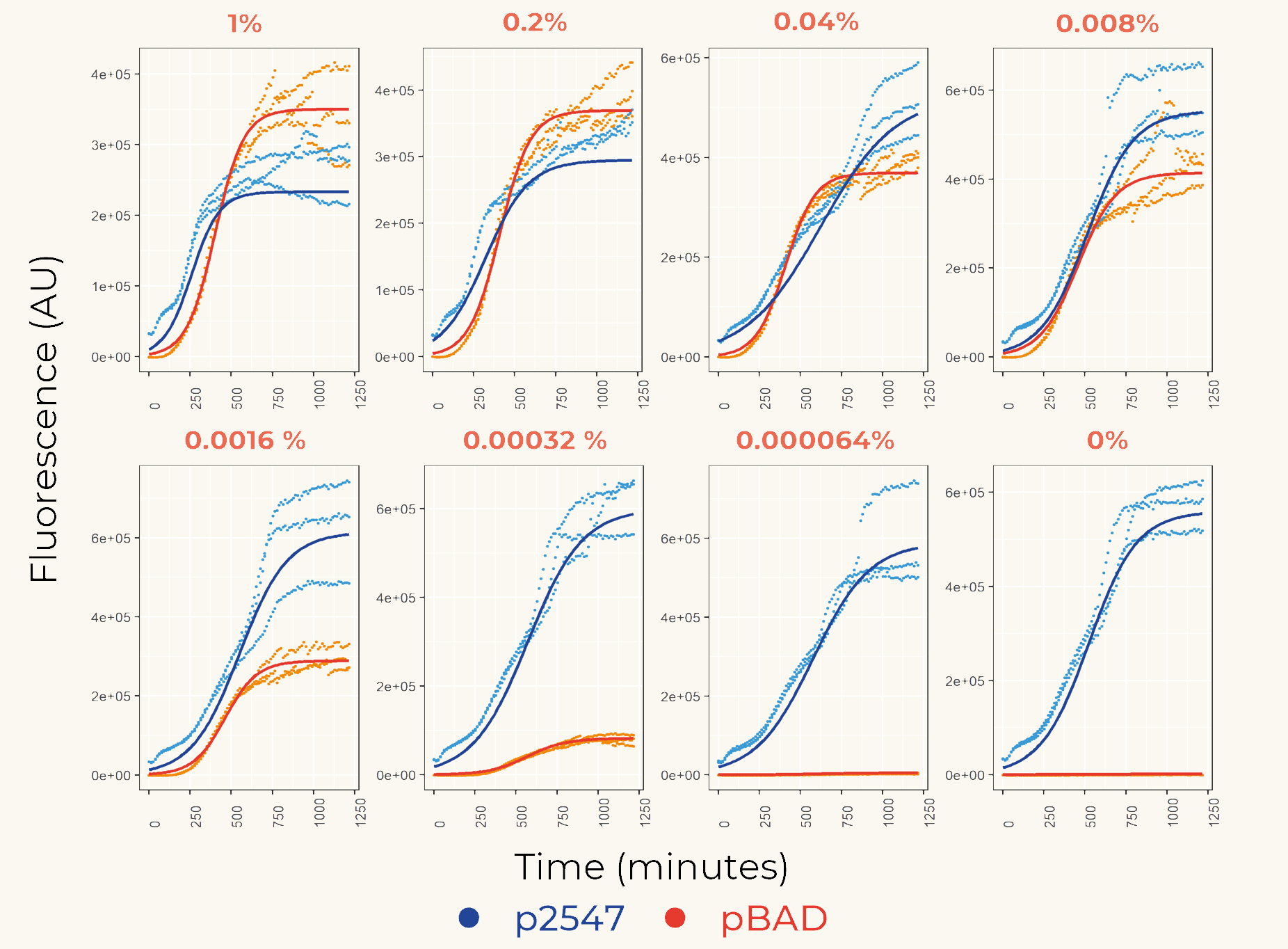
| + | We first cloned promoters in front of the gene of the fluorescent protein mCherry ([https://parts.igem.org/Part:BBa_K3962339 BBa_K3962339] and [https://parts.igem.org/Part:BBa_K3962340 BBa_K3962340]) for the calibration. The result showed constant expression from p2547::mCherry and the expression from pBAD::mCherry increased gradually as the concentration of arabinose increased (Figure 1). The lowest fluorescent protein expression was observed at the concentration of 0.00032% (w/v). We also concluded that pBAD is tightly regulated as it showed low leaky expression when no arabinose was added. | ||
| + | |||
| + | [[Image:T-Leiden-parts-Figuer1.png|500px]] | ||
| + | |||
| + | '''''Figure 1.''''' ''The fluorescence intensity of strains carrying our construct under different arabinose concentration. The measurement was carried out for 20 h. The x-axis shows the time of each measurement. The y-axis shows the fluorescence of mCherry in the arbitrary unit (AU). The blue data points represent the fluorescent intensity of p2547::mCherry strain, red data points represent the pBAD::mCherry strain. The arabinose concentration is shown at the top of each graph. '' | ||
| + | |||
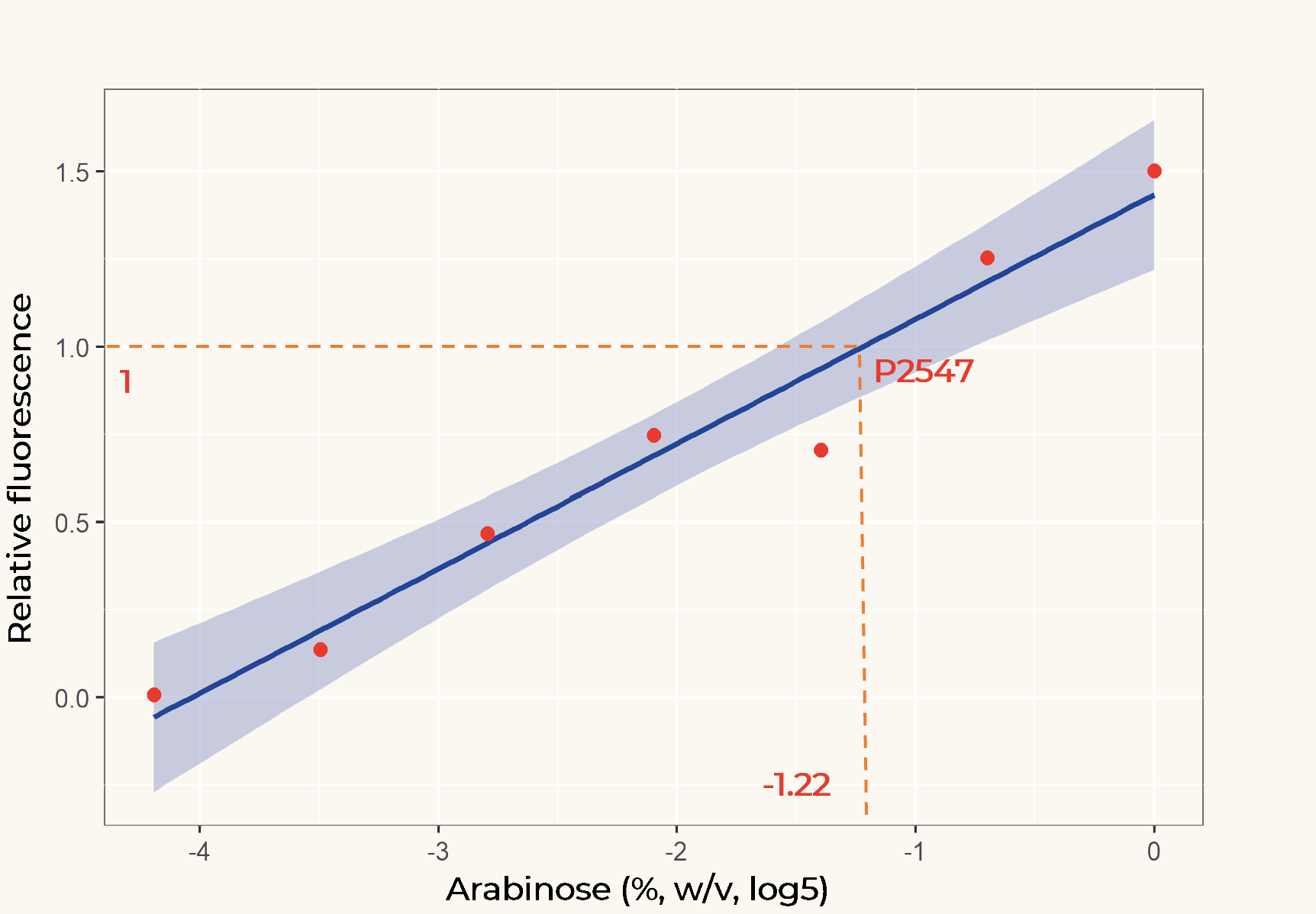
| + | The relation between the arabinose concentration (w/v, log5) and the relative fluorescence of pBAD fitted into a linear regression model with R2=0.9586 (Figure 2). From the regression it could be calculated that at the arabinose concentration of 0.14%, the transcriptional activity of pBAD was equal to p2547. | ||
| + | |||
| + | [[Image:T-Leiden-parts-Figuer2.png|500px]] | ||
| + | |||
| + | '''''Figure 2.''''' ''Linear regression of arabinose concentration relative fluorescence of pBAD compared with p2547. The orange dash lines showed how p2547 related to pBAD transcriptional activity at the arabinose concentration of 0.14% (log5-1.22).'' | ||
| + | |||
| + | To assist future iGEM teams in the usage of these promoters for regulation and prediction of synthetic circuitry, a table with arabinose concentrations causing equal expression of promoters in Anderson’s constitutive promoter collection was made. | ||
| + | |||
| + | [[Media:Anderson promoter family-pBAD.xls|Anderson promoter family-pBAD]] | ||
| + | |||
| + | |||
| + | = Advice = | ||
| + | |||
| + | We advice to use a consequent addition of arabinose in a low concentration if you want to have a high expression of the protein. | ||
| + | As this is very labour intensive, we advice to use another vector if you want to have a high expression of a certain protein. We therefore advise the usage of pET28a, which is induced with IPTG. | ||
| + | |||
| + | The reason we choose to use pBAD was because we planned to do a double transformation, which required different antibiotic resistance and different promoters. | ||
| + | |||
| + | ===Induction and Subsequent Inhibition of the pBAD Promoter=== | ||
| + | (Characterized by SDU-Denmark 2017)<br> | ||
| + | <b>Expression by the pBAD promoter can be regulated tightly by induction and subsequent inhibition.</b><br> | ||
| + | The pBAD promoter holds great potential to control the expression of genes that require tight regulation, as it is capable of both an induction and repression. The [http://2015.igem.org/Team:HKUST-Rice, HKUST-Rice iGEM team] from 2015 found that the pBAD promoter exhibits an almost all-or-none behaviour upon induction with arabinose when located on a high copy vector, but allows for gradual induction when cloned into a low copy vector. Thus, it was evident that this promoter on a high copy vector would be inappropriate for tightly regulated gene expression. Based on these findings, a low copy vector was used to investigate the ability to inhibit gene expression subsequent to induction of pBAD. <br> | ||
| + | |||
| + | Gene expression was simulated by fluorescence microscopy using a pBAD-YFP reporter system, [https://parts.igem.org/Part:BBa_I6058 BBa_I6058]. | ||
| + | For this purpose, an Olympus IX83 with a photometrics prime camera was used with an exposure time for YFP at 200 ms. Transformed <i>E. coli</i> MG1655 cells were cultured in M9 minimal medium supplemented with 0.2% glycerol and 30 µg/mL chloramphenicol, to avoid catabolite repression from glucose residues present in LB medium. Two cultures were incubated, of which one was induced with 0.2 % arabinose from the beginning. At OD<sub>600</sub>=0.1, designated time 0, the cultures were split in two and 0.2 % glucose was added to one of each pair. Samples were obtained at time 0, before division of the cultures, and at 30 min, 60 min, and 120 min. | ||
| + | The resulting images revealed, that the inducer arabinose was required to stimulate expression of YFP, and that the addition of the repressor glucose to a uninduced culture had no effect. Furthermore, it was evident that addition of arabinose induced expression of YFP, and that subsequent addition of glucose terminated the pBAD regulated gene expression, resulting in a reduced fluorescence level. 30 minutes after inhibition this reduction was already evident, and after 120 minutes the gene expression controlled by pBAD was even further decreased, as seen in Figure 1.<br> | ||
| + | |||
| + | <center> | ||
| + | https://static.igem.org/mediawiki/2017/archive/f/f5/20171102025131%21T--SDU-Denmark--pBAD-promoter.png | ||
| + | </center> | ||
| + | |||
| + | Figure 1. YFP fluorescence levels in <i>E. coli</i> MG1655 transformed with the pBAD-YFP reporter system on pSB3K3. Left: Cultures with the inducer arabinose added. Right: Cultures not induced with arabinose. Both cultures were split up at OD<sub>600</sub>=0.1, designated time 0, and the inhibitor glucose was added to one half of each culture. Images were obtained at 0, 30, 60, and 120 minutes.<br> | ||
| + | |||
| + | This experiment made it clear, that gene expression controlled by the pBAD promoter is both inducible and repressible as required when cloned into the low copy vector pSB3K3. | ||
| + | |||
| + | = Characterization by BNU-China 2022 = | ||
| + | https://2022.igem.wiki/bnu-china/<br> | ||
| + | |||
| + | This part contains an arabinose operon, which constitutes the arabinose sensing system. We added the ccdB gene downstream of the pBAD promoter which allows for CcdB protein expression in the presence of arabinose involving poisoning of DNA-topoisomerase II complexes. | ||
| + | <center>[[File:T--BNU-China--suicide_pathway.png|800px]]</center> | ||
| + | <center>Fig.1 The arabinose to specifically turn on the kill switch in engineered bacteria</center> | ||
| + | |||
| + | |||
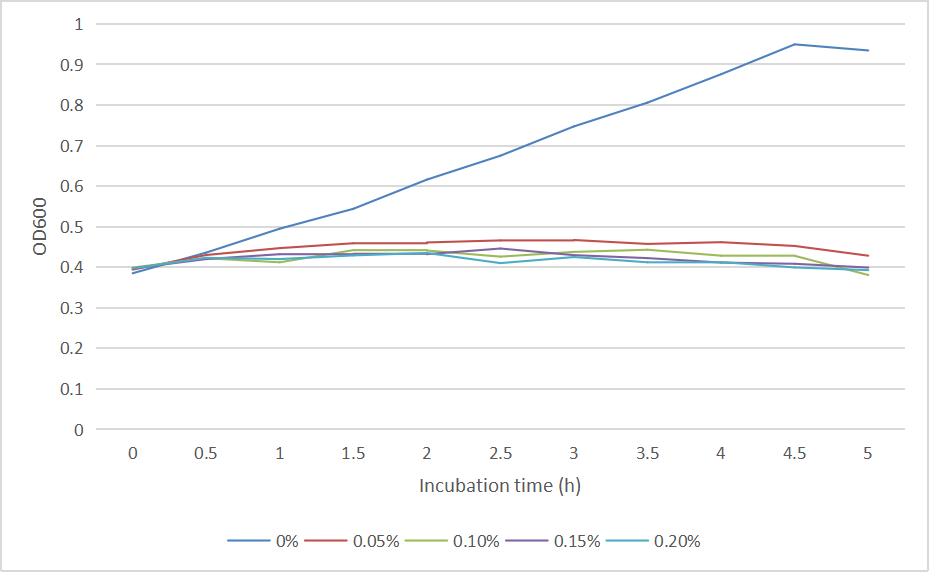
| + | We measured the OD600 of each sample to measure intact bacteria counts. We cultured the strain at 37°C overnight, obtained the bacteria by centrifugation, resuspended the bacteria in centrifuge tubes containing LB medium with different concentrations of arabinose (0%, 0.05%, 0.10%, 0.15%, 0.20%). We then added the resuspended bacteria to the corresponding concentration of arabinose medium to make a final volume of 20 mL, and measured the initial OD600. Samples were taken once every hour to measure OD600. And the diluted bacterial solution was spread on solid LB-kanamycin medium, incubated at 37°C overnight for the colony counting.<br> | ||
| + | |||
| + | The OD600 of the colonies is shown as below in '''Fig. 2''':<br> | ||
| + | |||
| + | <center>[[File:T--BNU-China--OD6001.png|600px]]</center><br> | ||
| + | |||
| + | <center>Fig.2 The OD600 of ''E. coli'' DH5α under arabinose</center><br> | ||
| + | |||
| + | The growth of colonies is shown in '''Fig. 3''':<br> | ||
| + | |||
| + | <center>[[File:T--BNU-China--colonies1.png|600px]]</center><br> | ||
| + | |||
| + | <center>Fig.3 Comparison of growth of the engineered bacteria treated with L-arabinose at different concentrations and time duration</center> | ||
| + | <center>(from left to the right: 1h, 2h, 3h, 4h; top to bottom: 0%, 0.05%, 0.1%, 0.15%, 0.2%)</center><br> | ||
| + | |||
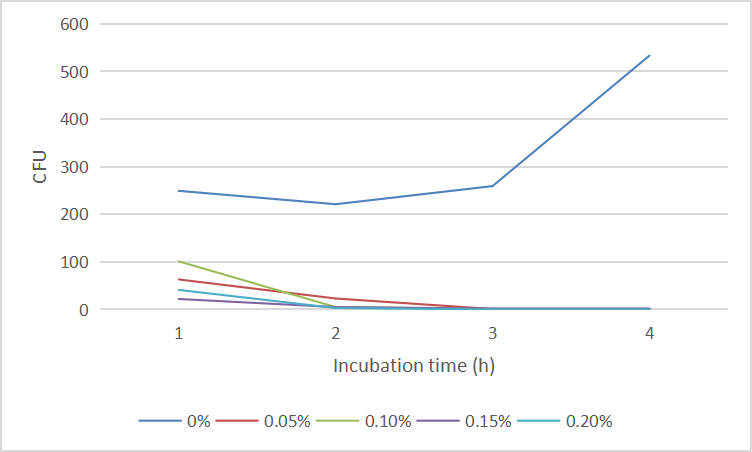
| + | And the cell number was concluded as CFU and graphed in '''Fig. 4''':<br> | ||
| + | |||
| + | <center>[[File:T--BNU-China--cell_number.png|600px]]</center><br> | ||
| + | |||
| + | <center>Fig.4 The CFU of ''E. coli'' DH5α under arabinose</center><br> | ||
| + | |||
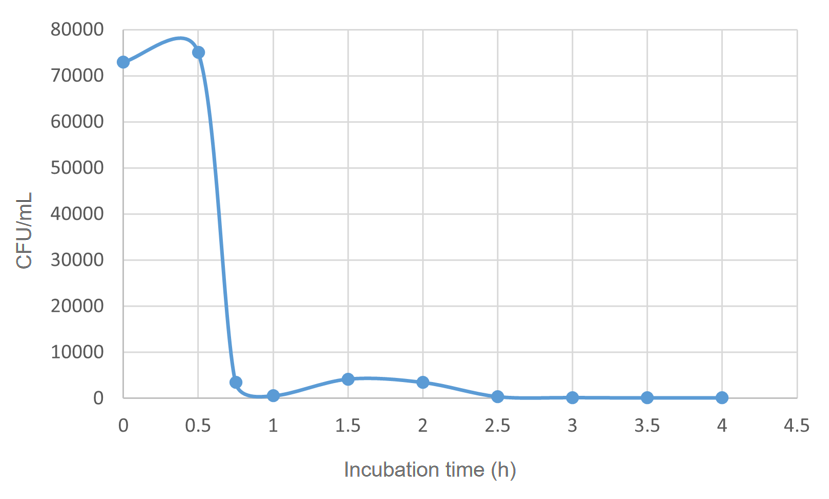
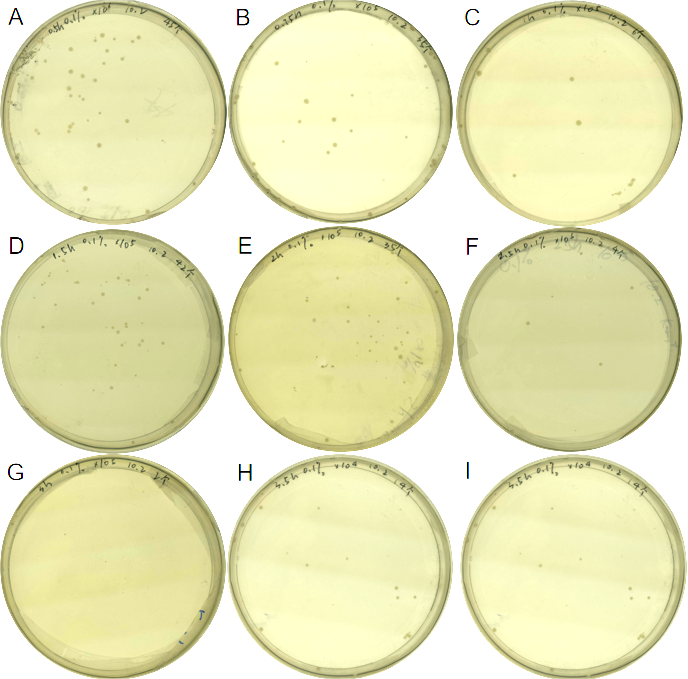
| + | We found that 0.1% concentration of arabinose was the most effective in killing cell, so we measured the effect of DH5α suicide induced by 0.1% arabinose. We took samples every 15 minutes at 0.5-1 hours when the bacterial concentration was rapidly decreasing, and every half hour for the rest of the time. The samples were diluted and spread on solid medium and counted for colonies, then we have the colonies shown in '''Fig. 5'''.<br> | ||
| + | |||
| + | <center>[[File:T--BNU-China--colonies01.png|600px]]</center><br> | ||
| + | |||
| + | <center>Fig.5 Comparison of growth of the engineered bacteria treated with 0.1% L-arabinose at different time duration, the dilution of the bacterial solution is also different</center> | ||
| + | <center> (A) 0.5h, 10<sup>6</sup> (B) 0.75h, 10<sup>5</sup> (C) 1h,10<sup>5</sup> (D) 1.5h, 10<sup>5</sup> (E) 2h, 10<sup>5</sup> (F) 2.5h, 10<sup>5</sup> (G) 3h,10<sup>5</sup> (H) 3.5h,10<sup>4</sup> (I) 4h, 10<sup>3</sup></center><br> | ||
| + | |||
| + | The colony numbers were collected and shown in '''Fig.6''':<br> | ||
| + | |||
| + | <center>[[File:T--BNU-China--cell_number01.png|600px]]</center><br> | ||
| + | |||
| + | <center>Fig.6 The CFU of ''E. coli'' DH5α under 0.1% arabinose</center><br> | ||
Latest revision as of 06:13, 12 October 2022
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_I0500
- When grown with 0.2% arabinose, promoter is weak-medium. [jb, 5/24/04] Part may not be compatible with MC4100 as cell line is araD 139.
- MC4100 is not a good chassis for operating BBa_I0500 (pBad promoter). The feed-forward regulation of the endogenous promoter controlling expression of the arabinose transporter prevents linear induction with increasing arabinose concentration. (Engineered strain from Keasling's lab, used by jrk for operation of the screening plasmid.) [cconboy 04]
- Observations of induced expression of GFP (BBa_E0840) are consistent with previous comments about weak promoter signal induction by jb 5/24/04. [melissali, Berkeley iGEM 2005]
PC and AraC are located on the complementary strand, reading right to left as written.
- At least one registry stock contains a deletion of the C at base 1194. This is after the transcriptional start but before the translation start, so it may not be significant. Parts with this mutation have been qualitatively observed to function normally.
- Can with success be combined with a promoter from pBAD SPL to obtain a very low leakiness and an appropriate strength. This is very efficient for expressing lethal proteins.
- Can be used for diauxic shifts with appropriate media as the promoter has a CAP binding site and thus shows catabolite repression with glucose [IISc Bangalore, 2016].
User Reviews
UNIQ4524817ca65a7cdc-partinfo-00000000-QINU
|
•••••
Antiquity |
This review comes from the old result system and indicates that this part worked in some test. |
|
iGEM Groningen 2009 |
The sequence was listed as inconsistent, and the ligations of parts behind the promoter failed. Restriction of isolated plasmids showed fragments of unexpected sizes. |
|
•••••
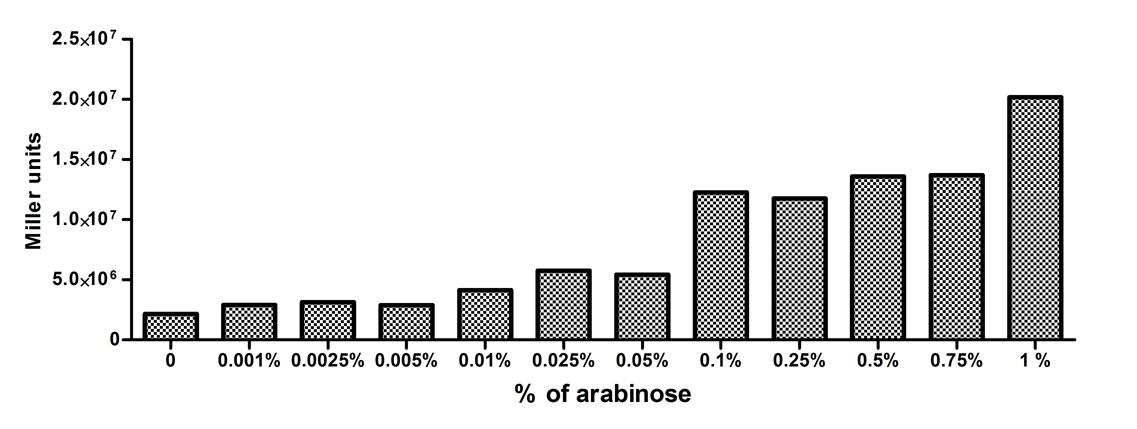
[http://2010.igem.org/Team:Slovenia 2010 iGEM team Slovenia] |
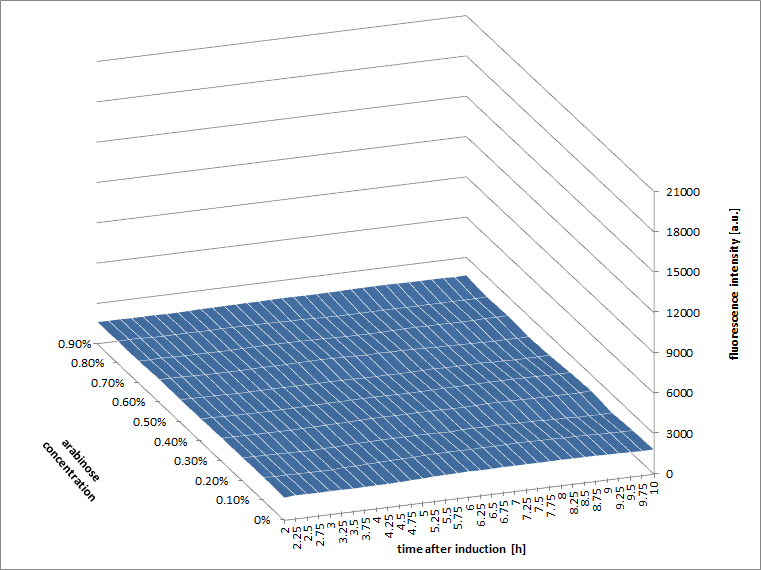
[http://2010.igem.org/Team:Slovenia 2010 iGEM team Slovenia] further characterized the pBAD/araC promoter (Part:Bba_I0500) using lacZ gene that encodes for beta-galactosidaze enzyme (Part:Bba_K323133). pBAD promoter is an arabinose inducible promoter. When incubating E.coli cultures containing the lacZ under pBAD/araC promoter, increasing concentrations of arabinose leads to higer promoter activity that results in higher amounts and subsequently higer activity of beta-galactosidase enzyme as seen from figure below. The resulting graph shows a trend of increasing beta-galactosidase activity over increasing concentrations of arabinose. The tested cultures incubated at different concentrations of arabinose (i.e. 0%, 0.001%, 0.0025%, 0.005%, 0.01%, 0.025%, 0.05%, 0.1%, 0.25%, 0.5%, 0.75% and 1%) have shown a trend of increasing beta-galactosidase activity with raising arabinose concentration. The activity was highest at 1% added arabinose, which implies the promotor should be induced by this arabinose concentration for reaching maximum expression of the regulated protein. However, some beta-galactosidase activity was observed even at 0% added arabinose, which indicates slight leakage of the pBAD promoter. pBAD/araC_lacZ_DTER construct (Part:Bba_K323133) includes a beta-galactosidase (lacZ) gene, the expression of which is regulated by the pBAD promoter (Part:Bba_I0500). This construct was designed for the sole purpose of characterising the pBAD promoter with the beta-galactosidase assay (for a detailed description of the method see the 2010 iGEM team Slovenia wiki). |
|
•••••
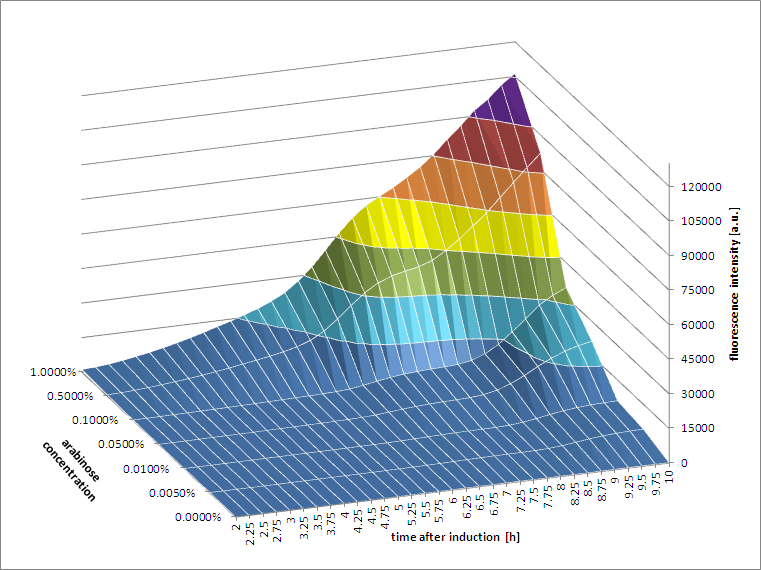
[http://2011.igem.org/Team:Groningen 2011 iGEM team Groningen] |
[http://2011.igem.org/Team:Groningen 2011 iGEM team Groningen] characterized the pBAD/AraC promoter (Part:Bba_I0500) even further using GFP and measuring the fluorescence in a fluorimeter over time, utilizing different arabinose concentrations to induce the promoter. After analysis of the obtained data (see graphs below), it is clear that the induction is faster and stronger with higher arabinose concentrations. It shows a dynamic response over a range of arabinose concentrations and over time. Because of that it can be treated as a precise tool capable of giving different output levels. The measurements were done in DH5alpha strain carrying a pSB1C3 with BBa_K607036 in triplicate.
|
|
•••••
[http://2011.igem.org/Team:Cambridge Cambridge 2011] |
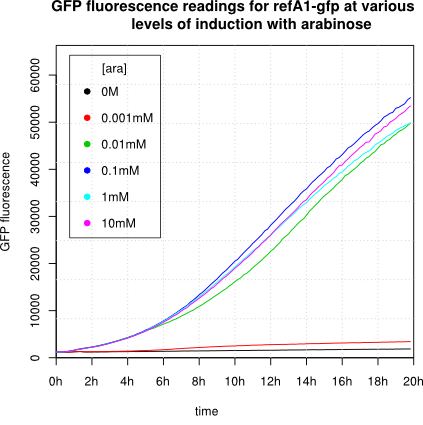
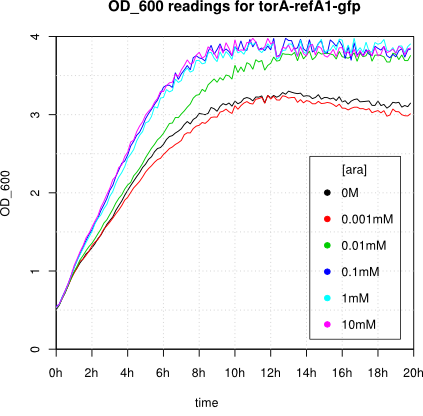
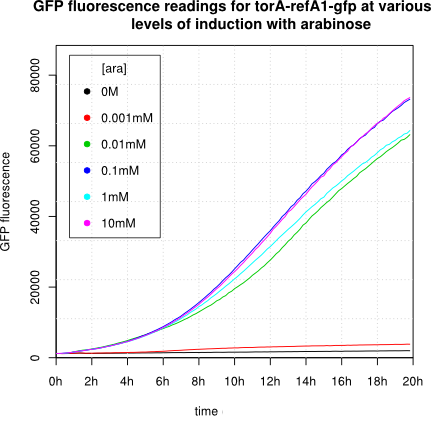
We observed an 'all-or-nothing' response in our use of the pBAD promoter. We grew up cells of the strain [http://cgsc.biology.yale.edu/Strain.php?ID=111773 BW27783] overnight at 37°C. We then diluted the culture 10-fold and grew the cells back up to an OD_600 of 0.5 to catch the cells in mid-exponential phase. We then induced with arabinose at various concentrations from 0.001mM up to 10mM and took OD and GFP readings on a plate reader over the next 20 hours. During this time the cells were held at 37°rees;C. The cells contained our ReflectinA1-sfGFP and TorA-ReflectinA1-sfGFP constructs on pSB3K3 plasmids. This work was not intended to characterise the pBAD promoter, but it highlights some of the difficulties in doing so:
Note: 1mM arabinose corresponds to 0.015% w/v.
|
|
[http://2012.igem.org/Team:UNITN-Trento Trento 2012] |
We were unable to extract this part from the distribution kit, so we developed our own! Please head to BBa_K731201 for documentation and characterization. |
|
••••
[http://2014.igem.org/Team: Hong_Kong_HKUST 2014] |
2014 HKUST iGEM team previously submitted [http://2014.igem.org/Team:Hong_Kong_HKUST/riboregulator/characterization characterization data] of Part BBa_I0500 that described an all-or-none behavior of the promoter. That is however not the case. We sincerely apologize for the misleading information. Please refer to our latest characterization data by HKUST-Rice 2015 below instead. UNIQ4524817ca65a7cdc-partinfo-0000000A-QINU |
|
•••••
[http://2015.igem.org/Team: HKUST-Rice 2015] |
[http://2015.igem.org/Team:HKUST-Rice HKUST-Rice Team] characterized BBa_I0500 using the construct BBa_I13540 in low copy plasmid pSB3K3. We aimed to obtain a dose response curve capturing the dynamic range and maximum induction concentration in order to compare its behavior when it is [http://2015.igem.org/Team:HKUST-Rice/Expression expressed in parallel with another sensor]. (please see BBa_K1682017 for more details. 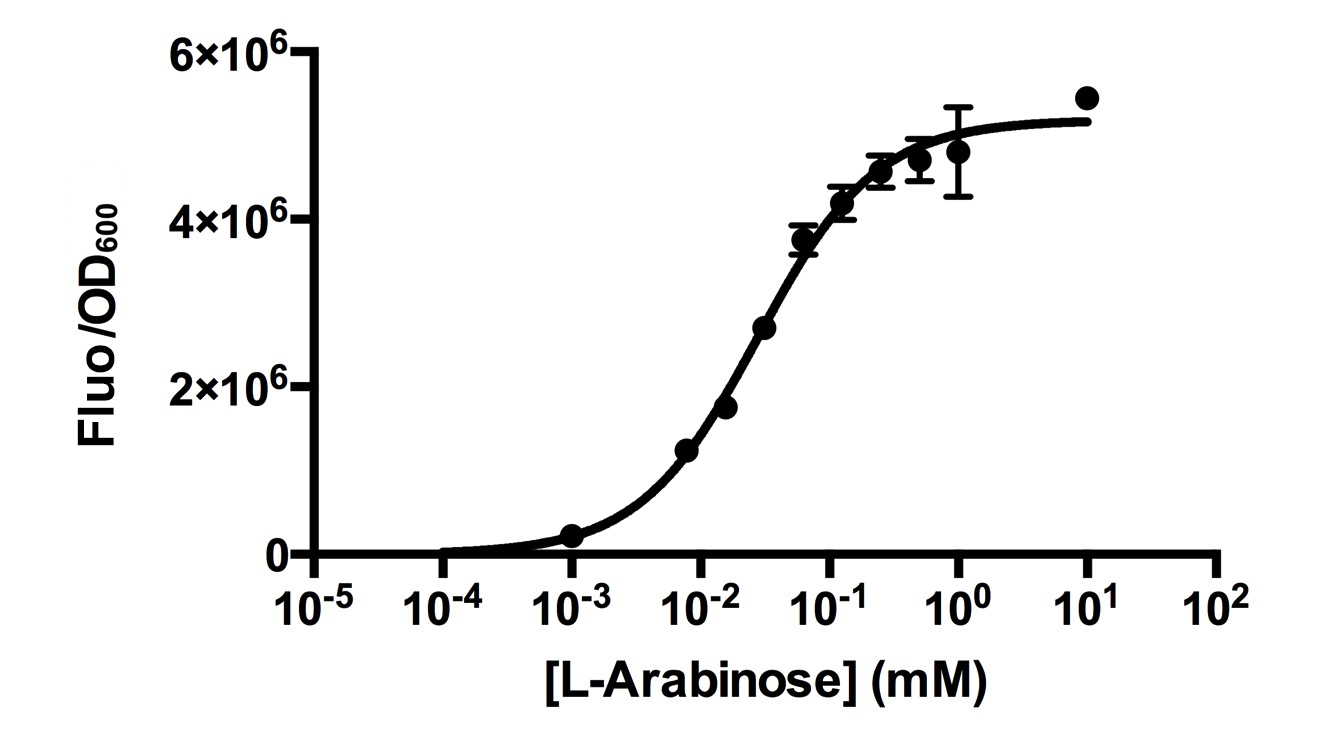 Figure 1. Dose response curve of cells carrying pSB3K3-BBa_I13540 under increasing L-arabinose concentration. Fluo = Relative Fluorescence Units. Error bars represent SD (n=3). Results indicate that the sensing range of BBa_I0500 lies at roughly 10-3 to 10-1 mM and has a half-maximal induction concentration of around 3-2mM of Arabinose.
Our results demonstrated that this promoter has a graded induction response and different sensing ranges on plasmids with different copy numbers (Figure 2). In concordance with literature, it displays an all-or-none behavior on a single cell level when expressed from a high copy plasmid pSB1K3, but not when expressed from a low copy plasmid pSB3K3 (Figure 3). 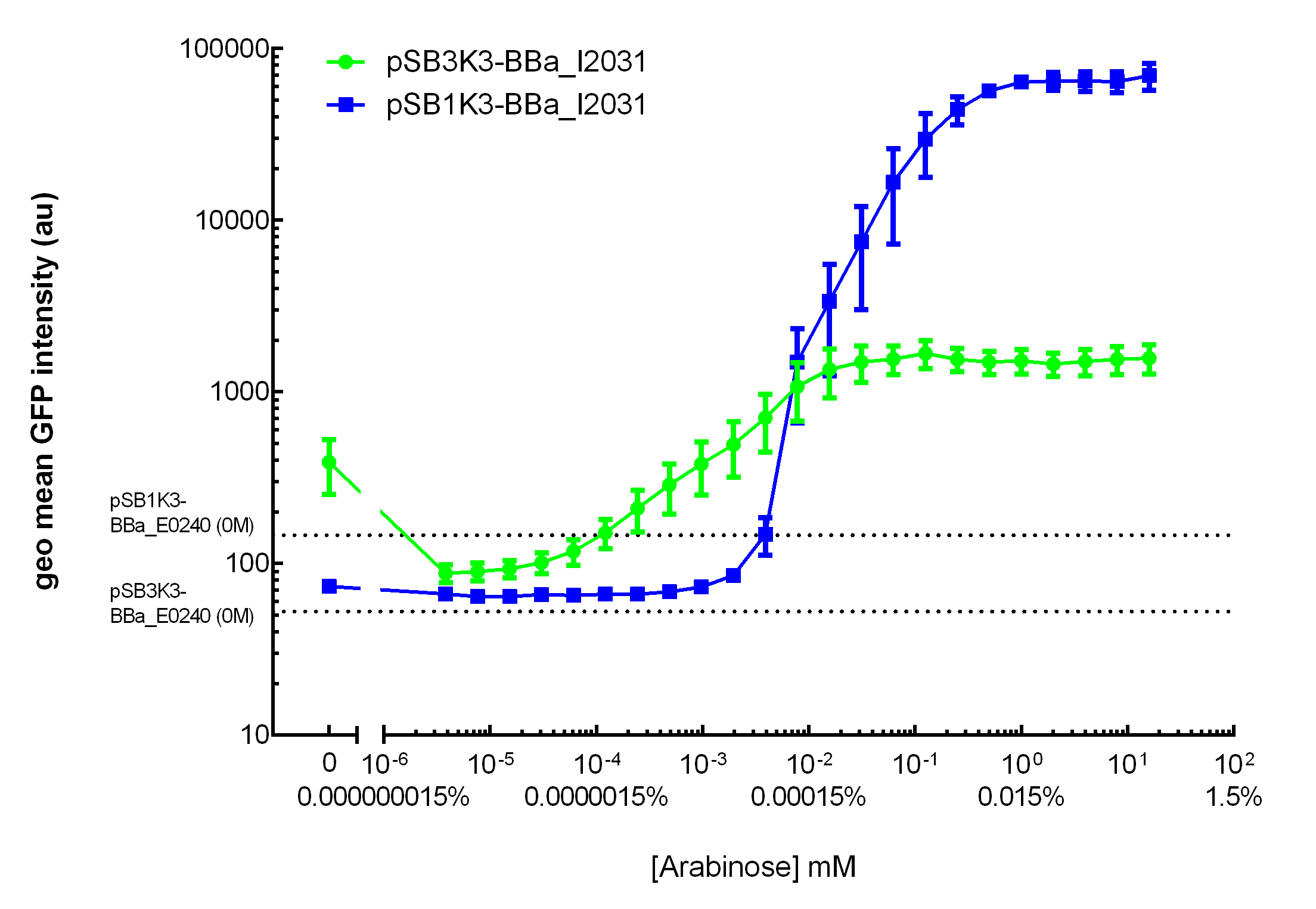 Figure 2. Transfer curves for BBa_I2031 on plasmid pSB3K3 and pSB1K3. On pSB3K3, BBa_I0500 is responsive to 10-4 - 10-2 mM arabinose, whereas on pSB1K3, it senses arabinose from roughly 10-3 to 1mM. These data agreed with neither Cambridge 2011 nor Groningen 2011’s result. The dashed lines represent cells’ auto fluorescence, measured using DH10B cells harboring pSB3K3-BBa_E0240 or pSB1K3-BBa_E0240. Error bars represent SEM of 3 independent experiments on 3 different days. The molar concentration to percentage conversion was provided for comparison with previous data. 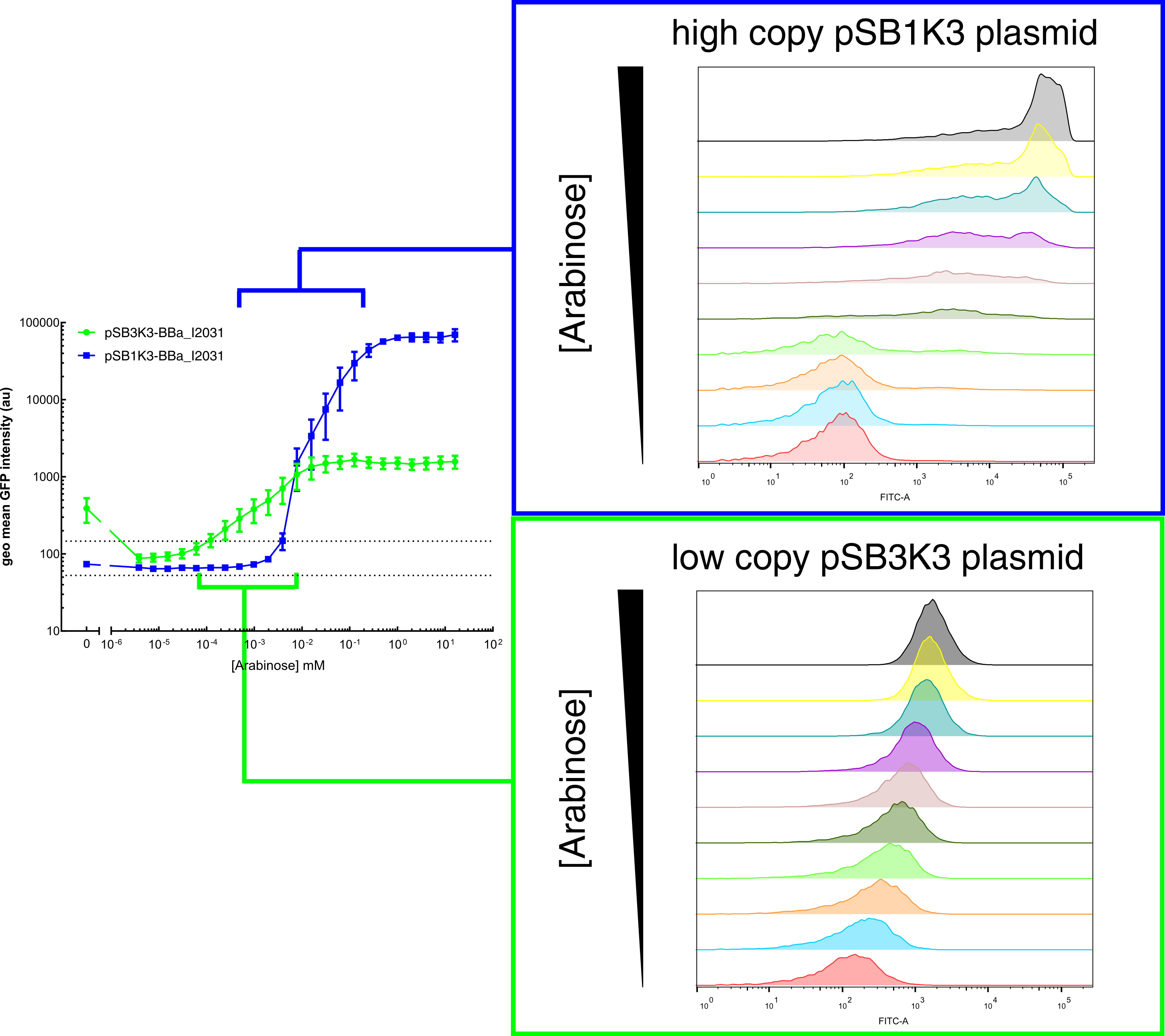 Figure 3. Histogram plots for sensing ranges of BBa_I0500 on high and low copy plasmid. An all-or-none induction can be observed when BBa_I0500 is placed on a high copy plasmid, where induced cells were mostly distributed among two bins of fluorescence. Yet on low copy plasmid, the induced populations remain homogenous along the arabinose concentration gradient. Concentrations of arabinose for high copy pSB1K3 plasmid: 0.488µM – 0.25mM. For low copy pSB3K3 plasmid: 0.0610µM – 0.03125mM. Only 1 set of experiment result from 3 replicates is presented. Moreover, we would like to warn potential users of this part not to drive expression of riboregulators ([http://www.ncbi.nlm.nih.gov/pubmed/15208640 Isaacs et al., 2004]) using BBa_I0500. This PBAD promoter contains a 19bp sequence after the transcription start site, which would produce a taRNA with additional nucleotides at its 5’ end, and rendering it incapable to activate crRNA. To avoid misuse of this part due to incognizance of its sequence information, we recommend users to look up BBa_K1067007 for annotations. For more information and methods of our characterization, please refer to our detailed characterization report available through our [http://2015.igem.org/Team:HKUST-Rice/Expression/ParaBAD wiki page]. |
|
•••••
[http://2016.igem.org/Team:IISc_Bangalore]Team IISc Bangalore |
IISc Bangalore characterized BBa_I0500 by using Bba_746908 which has sfGFP under the pBAD/araC promoter. We wanted to verify the catabolite repression of the expression from the promoter in the presence of glucose. |
|
•••••
[http://2016.igem.org/Team:OUC-China]Team OUC-China |
[http://2016.igem.org/Team:OUC-China OUC-China] characterized BBa_I0500 of different concentrations of L-arabinose on the transcriptional level. We found that the strength of I0500 was mainly measured on the translational level before. As far as we are concerned, it is the transcriptional level that can directly reflect I0500’s original strength without interfering by context. In order to verify it more accurately, we tested it on the transcriptional level by extracting RNA and measured it by qPCR. We constructed I0500 with GFP(E0040) and transformed into TOP10F’. Firstly , we plated on LB agar with chloramphenicol and individual colonies were subsequently grown overnight in LB-medium. Cultures were back diluted 1:100 into 100mL fresh LB-medium with chloramphenicol, adding 0,0.0002%,0.002%,0.02%,0.2% (m/V)L-arabinose. After growing for 18 hours, we extracted RNA and measured it. At the same time, we measured the fluorescence of GFP(485nm/520nm). Besides, we also measured OD over time to check whether L-arabinose promotes bacteria’s growth. 1. 100mlLB,blank |
|
••••
[http://2017.igem.org/Team:Glasgow]Team Glasgow 2017 |
[http://2017.igem.org/Team:Glasgow Team Glasgow 2017] improved this part by splitting its constituent parts into two separate BioBricks: BBa_K2442101 encoding minimal pBAD and BBa_K2442104 encoding AraC under regulation of LacI-regulated promoter. This allows for greater control over arabinose-inducible systems. Placing AraC under control of LacI regulated promoter (R0011) allows to induce expression of AraC only when required, taking translational load off grown cells. R0011 is inducible by IPTG. AraC coding sequence alone is also available as part BBa_K2442103, which can be placed under other promoter of choice. We isolated the minimal DNA sequence from the native pBAD that is sufficient to retain its function. The promoter retains all the operator sites O1, O2, I1 and I2 required for binding of AraC. As some of these sites lie within the PC promoter and overlap with AraC coding sequence, the minimal pBAD has the araC start codon ATG mutated to AGT to prevent the protein from being made. See part BBa_K2442101 and our [http://2017.igem.org/Team:Glasgow/araC wiki] for details. We tested pBAD with the use of pBAD+GFP reporter plasmid (BBa_K2442102) by measuring fluorescence. Activity of the promoter was tested in presence of arabinose, xylose, decanal and no inducer (Fig. 1). When transformed into AraC-positive E. coli strains such as DH5α, pBAD is sufficiently activated with just the chromosomal copy of araC and does not require introduction of araC from external plasmid. Moreover, DH5α cells carrying K2442102 showed fluorescence in presence of arabinose, but no significant increase in fluorescence in other conditions, showing high specificity of AraC and minimal leakage of minimal pBAD. DH5α carrying pBAD+GFP reporter on a high copy number plasmid (pSB1C3) BBa_K2442102 showed 2000x fold increase in fluorescence on arabinose plate compared to empty cells (Fig. 1). This proves significant improvement of part BBa_I0500. DH5α cells carrying minimal pBAD and I13500 on a low copy number plasmid pSB3K3 instead of pSB1C3 also showed expression of GFP only under arabinose conditions. However, levels of fluorescence were much lower, suggesting that the level of GFP production is dependent on the plasmid copy number and must be taken into account in further experiments. In AraC-negative strains, AraC expressed from BBa_K2442104 induces pBAD with similar efficiency. Overall, results show that both AraC and arabinose are necessary to activate pBAD. Efficiency of minimal pBAD is demonstrated by 2000x fold increase in fluorescence of cells carrying the reporter plasmid.  Figure 1:Average relative fluorescence over optical density at hour 8. Shows fluorescence levels under: no additive, 40mM Arabinose, 40mM Xylose and 2mM decanal. The E. coli strains which the plasmids have been transformed into are DS941 or DH5α (specified). DH5α is AraC-positive. DS941 is AraC-negative. BioBricks (K244210..) specified. All parts are in plasmid backbone pSB1C3 unless specified otherwise (pSB3k3). K2442102: reporter plasmid pBADmin+GFP. K2442104: regulatory plasmid R0011+B0032+AraC
|
|
•••••
BHSF_ND iGEM 2019 |
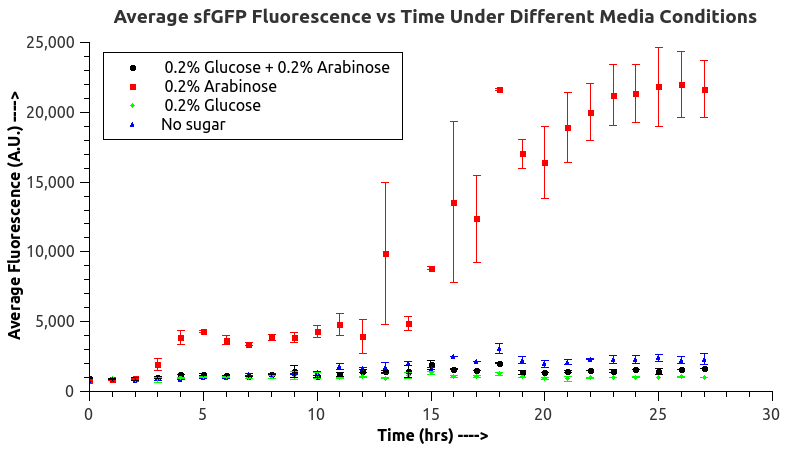
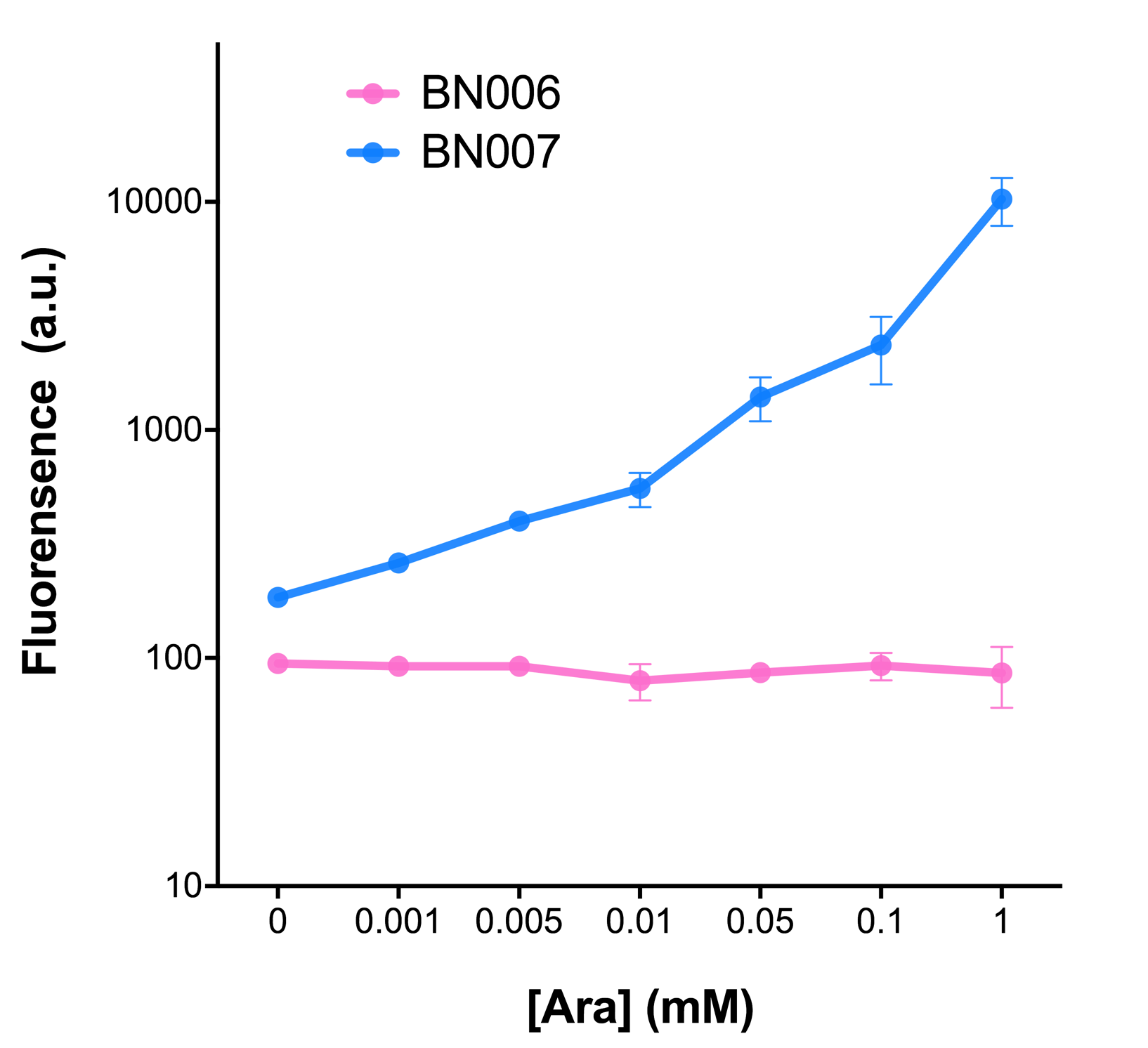
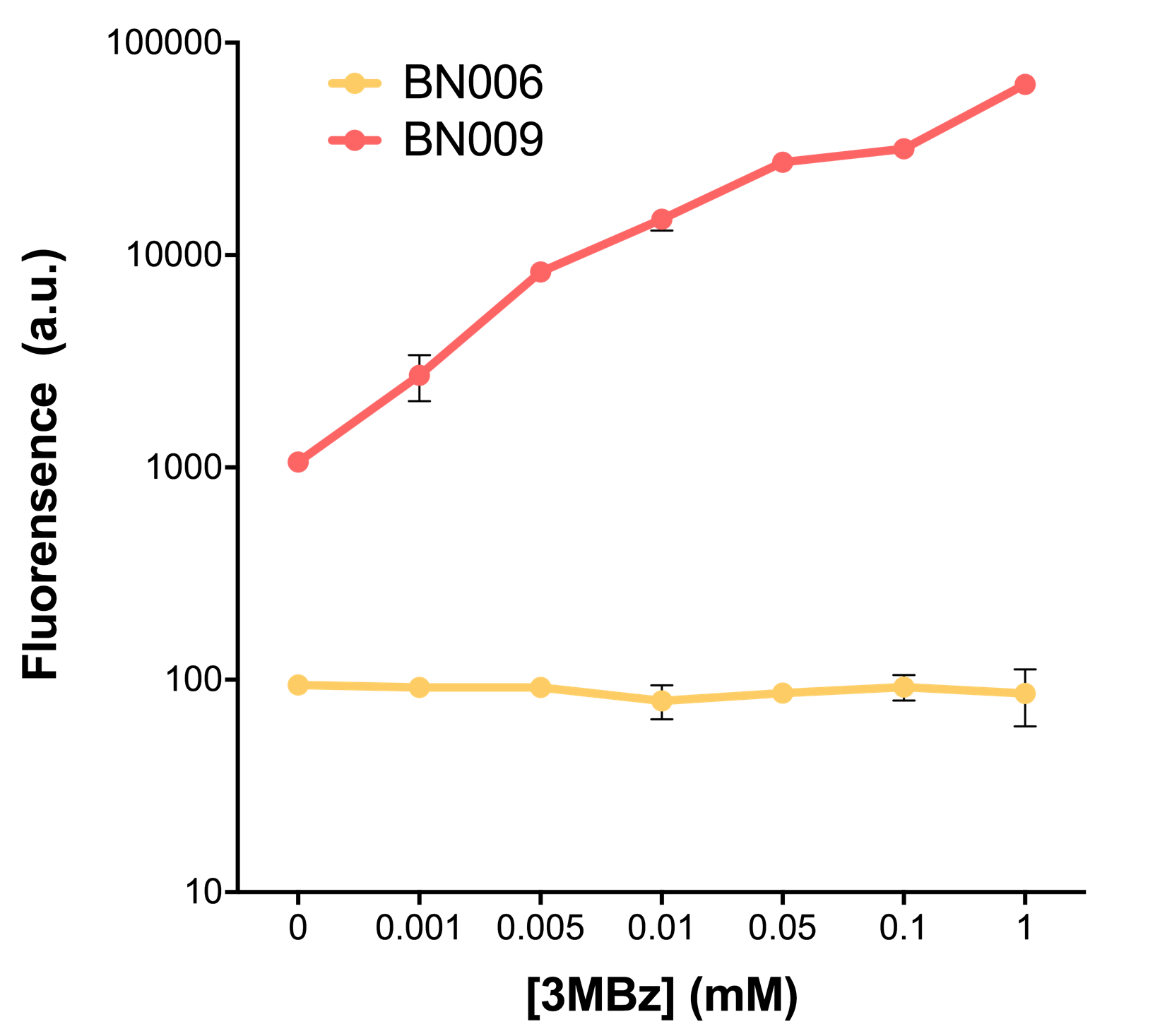
We mainly focused on two types of inducible transcriptional promoter—Xyls and pBAD. The Xyls protein is the positive transcription regulator of the TOL plasmid meta-cleavage pathway operon Pm. Xyls belongs to the AraC family of transcriptional regulators and exhibits an N-terminal domain involved in effector recognition, and a C-terminal domain.The activator Xyls is usually is usually activated by the benzoate and its derivatives (3MBz in our design). The pBAD promoter holds great potential to control the expression of genes that require tight regulation, as it is capable of both an induction and repression. The pBAD promoter exhibits an almost all-or-none behavior upon induction with arabinose when located on a high copy vector, but allows for gradual induction when cloned into a low copy vector. Based on these research findings, a low copy vector was used to investigate the ability to inhibit gene expression subsequent to induction of pBAD. As the result shows, both system exhibit relative high level of fluorescence compared to our backbone design( BN006/Contains BBa_K3202029) which suggest the two inducible transcription system function properly.
ResultsIn our experiment of leakage testing, we used low-copying plasmid backbone PSB4K5, with a PSC101 origin of replication. Two inducers with their respective promoters (AraC & pBAD; Xyls & Pm) are coupled with sfGFP to see if there is actually expression leakage when inducer is present quantitatively through flow cytometry. We tested fluorescence under different inducer concentration at the given time of 240 minutes and under different time stage at a given inducer concentration. Theoretically when inducers (AraC and 3MBz ) are present, promoters (pBAD and Pm) are initiated therefore both sfGFP are expressed; while sfGFP shouldn’t be expressed if inducer is absent. When measured at the end of 240 minutes without inducer, BN009/Contains BBa_K3202031(Xyls+sfGFP) has a basal leakage of 968 a.u. compared to BN006/Contains BBa_K3202029(backbone without GFP). BN007/Contains BBa_K3202030(pBAD+sfGFP) also showed a leakage of 90 a.u. The result suggested that both system have noticeable leakage and Xyls has much higher leakage. When we gradually increased inducer concentration to 1mM, the fluorescence of BN006/Contains BBa_K3202029 stays at a constant level, whereas the fluorescence of both Xyls and pBAD have increased exponentially. This is another sign that both system function properly. We found an intriguing result when measuring the fluorescence of the two system under a constant inducer concentration of 1mM. During the first 160 minutes, the fluorescence of BN007/Contains BBa_K3202030 is 200 a.u. higher than BN006/Contains BBa_K3202029. However, at 240 minutes, the fluorescence value suddenly increased to 7641 a.u., which is 7590 a.u., higher than the backbone. Same thing happened to BN009/Contains BBa_K3202031. Therefore, in the subsequent experiments, systems are induced to up to 4 hours to ensure the accuracy of the experiment. Results shown above verified the presence of expression leakage of both systems when inducer is not expressed through the detectable fluorescence of reporter protein using flow cytometry. |
Contribution iGEM TU-Eindhoven 2017 [http://2017.igem.org/Team:TU-Eindhoven]
Method
We first tested three different temperatures, being 20, 25 and 37 degree Celsius. From our fist basic measurements, we discovered that expression was more efficient at a temperature of 37 degree Celsius and continued with this setting on our incubator. The next step was consequent adding arabinose to the cultures, as E. coli BL21(DE3) is a strain that can break down arabinose, and for an optimal expression, it is better to have a high enough constant concentration. We tested an addition of every 60 minutes and every 90 minutes, with varying concentrations of arabinose. The percentages that are depicted in the figures are the total concentration of arabinose in the whole culture. The indication half means that we added 50% of the original added amount of arabinose compared with the first addition. This was done because we expected that a higher initial dose was needed to induce the expression, but that not all added arabinose would be broken down, and we therefore didn't need to add a high amount of arabinose.
pBAD expression of fluorophore mCherry
The results of all our tries are depicted in Figure a1 & a2 with the best results in a3.
Figure a1: pBAD expression of mCherry in BL21(DE3) with every 60 minutes an addition of arabinose
Figure a2: pBAD expression of mCherry in BL21(DE3) with every 90 minutes an addition of arabinose
Figure a3: pBAD expression of mCHerry in BL21(DE3) most successful addition methods
pBAD expression of fluorophore GFP
The results of all our tries are depicted in Figure b1 & b2 with the best results in b3.
Figure b1: pBAD expression of GFP in BL21(DE3) with every 60 minutes an addition of arabinose
Figure b2: pBAD expression of GFP in BL21(DE3) with every 90 minutes an addition of arabinose
Figure b3: pBAD expression of GFP in BL21(DE3) most successful addition methods
pBAD expression conclusion
The best results are depicted in Figure c.
Figure c: pBAD expression of mCherry and GFP in BL21(DE3) most successful addition methods
2021 Team Leiden
https://2021.igem.org/Team:Leiden
We first cloned promoters in front of the gene of the fluorescent protein mCherry (BBa_K3962339 and BBa_K3962340) for the calibration. The result showed constant expression from p2547::mCherry and the expression from pBAD::mCherry increased gradually as the concentration of arabinose increased (Figure 1). The lowest fluorescent protein expression was observed at the concentration of 0.00032% (w/v). We also concluded that pBAD is tightly regulated as it showed low leaky expression when no arabinose was added.
Figure 1. The fluorescence intensity of strains carrying our construct under different arabinose concentration. The measurement was carried out for 20 h. The x-axis shows the time of each measurement. The y-axis shows the fluorescence of mCherry in the arbitrary unit (AU). The blue data points represent the fluorescent intensity of p2547::mCherry strain, red data points represent the pBAD::mCherry strain. The arabinose concentration is shown at the top of each graph.
The relation between the arabinose concentration (w/v, log5) and the relative fluorescence of pBAD fitted into a linear regression model with R2=0.9586 (Figure 2). From the regression it could be calculated that at the arabinose concentration of 0.14%, the transcriptional activity of pBAD was equal to p2547.
Figure 2. Linear regression of arabinose concentration relative fluorescence of pBAD compared with p2547. The orange dash lines showed how p2547 related to pBAD transcriptional activity at the arabinose concentration of 0.14% (log5-1.22).
To assist future iGEM teams in the usage of these promoters for regulation and prediction of synthetic circuitry, a table with arabinose concentrations causing equal expression of promoters in Anderson’s constitutive promoter collection was made.
Advice
We advice to use a consequent addition of arabinose in a low concentration if you want to have a high expression of the protein. As this is very labour intensive, we advice to use another vector if you want to have a high expression of a certain protein. We therefore advise the usage of pET28a, which is induced with IPTG.
The reason we choose to use pBAD was because we planned to do a double transformation, which required different antibiotic resistance and different promoters.
Induction and Subsequent Inhibition of the pBAD Promoter
(Characterized by SDU-Denmark 2017)
Expression by the pBAD promoter can be regulated tightly by induction and subsequent inhibition.
The pBAD promoter holds great potential to control the expression of genes that require tight regulation, as it is capable of both an induction and repression. The [http://2015.igem.org/Team:HKUST-Rice, HKUST-Rice iGEM team] from 2015 found that the pBAD promoter exhibits an almost all-or-none behaviour upon induction with arabinose when located on a high copy vector, but allows for gradual induction when cloned into a low copy vector. Thus, it was evident that this promoter on a high copy vector would be inappropriate for tightly regulated gene expression. Based on these findings, a low copy vector was used to investigate the ability to inhibit gene expression subsequent to induction of pBAD.
Gene expression was simulated by fluorescence microscopy using a pBAD-YFP reporter system, BBa_I6058.
For this purpose, an Olympus IX83 with a photometrics prime camera was used with an exposure time for YFP at 200 ms. Transformed E. coli MG1655 cells were cultured in M9 minimal medium supplemented with 0.2% glycerol and 30 µg/mL chloramphenicol, to avoid catabolite repression from glucose residues present in LB medium. Two cultures were incubated, of which one was induced with 0.2 % arabinose from the beginning. At OD600=0.1, designated time 0, the cultures were split in two and 0.2 % glucose was added to one of each pair. Samples were obtained at time 0, before division of the cultures, and at 30 min, 60 min, and 120 min.
The resulting images revealed, that the inducer arabinose was required to stimulate expression of YFP, and that the addition of the repressor glucose to a uninduced culture had no effect. Furthermore, it was evident that addition of arabinose induced expression of YFP, and that subsequent addition of glucose terminated the pBAD regulated gene expression, resulting in a reduced fluorescence level. 30 minutes after inhibition this reduction was already evident, and after 120 minutes the gene expression controlled by pBAD was even further decreased, as seen in Figure 1.

Figure 1. YFP fluorescence levels in E. coli MG1655 transformed with the pBAD-YFP reporter system on pSB3K3. Left: Cultures with the inducer arabinose added. Right: Cultures not induced with arabinose. Both cultures were split up at OD600=0.1, designated time 0, and the inhibitor glucose was added to one half of each culture. Images were obtained at 0, 30, 60, and 120 minutes.
This experiment made it clear, that gene expression controlled by the pBAD promoter is both inducible and repressible as required when cloned into the low copy vector pSB3K3.
Characterization by BNU-China 2022
https://2022.igem.wiki/bnu-china/
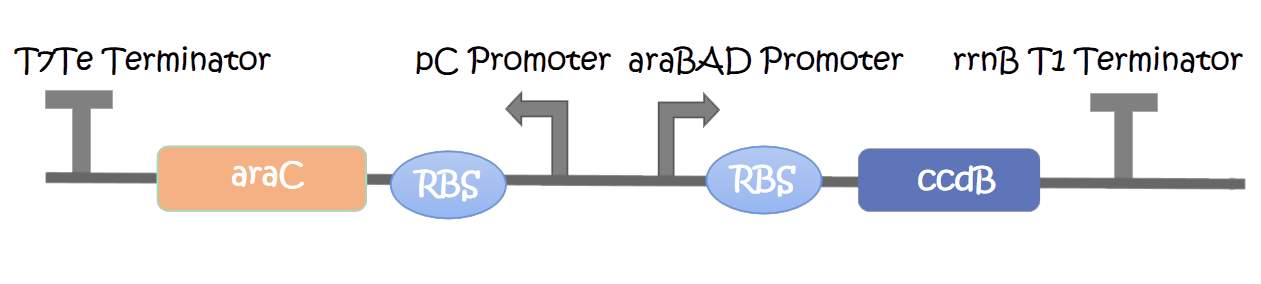
This part contains an arabinose operon, which constitutes the arabinose sensing system. We added the ccdB gene downstream of the pBAD promoter which allows for CcdB protein expression in the presence of arabinose involving poisoning of DNA-topoisomerase II complexes.
We measured the OD600 of each sample to measure intact bacteria counts. We cultured the strain at 37°C overnight, obtained the bacteria by centrifugation, resuspended the bacteria in centrifuge tubes containing LB medium with different concentrations of arabinose (0%, 0.05%, 0.10%, 0.15%, 0.20%). We then added the resuspended bacteria to the corresponding concentration of arabinose medium to make a final volume of 20 mL, and measured the initial OD600. Samples were taken once every hour to measure OD600. And the diluted bacterial solution was spread on solid LB-kanamycin medium, incubated at 37°C overnight for the colony counting.
The OD600 of the colonies is shown as below in Fig. 2:
The growth of colonies is shown in Fig. 3:
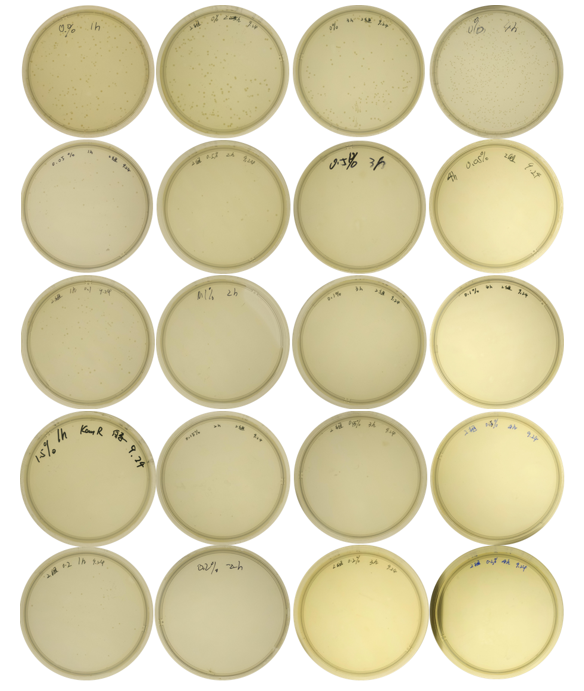
And the cell number was concluded as CFU and graphed in Fig. 4:
We found that 0.1% concentration of arabinose was the most effective in killing cell, so we measured the effect of DH5α suicide induced by 0.1% arabinose. We took samples every 15 minutes at 0.5-1 hours when the bacterial concentration was rapidly decreasing, and every half hour for the rest of the time. The samples were diluted and spread on solid medium and counted for colonies, then we have the colonies shown in Fig. 5.
The colony numbers were collected and shown in Fig.6: