Difference between revisions of "Part:BBa K1933001"
(→growth curve) |
|||
| (6 intermediate revisions by the same user not shown) | |||
| Line 21: | Line 21: | ||
==Characterization== | ==Characterization== | ||
| − | |||
| − | |||
''' Sequence confirmed! ''' | ''' Sequence confirmed! ''' | ||
| + | |||
| + | ''' <big><big>We further characterized functionality and improved [https://parts.igem.org/Part:BBa_K811005 BBa_K811005] from Penn 2012!!</big></big> ''' | ||
| + | |||
===Western blotting=== | ===Western blotting=== | ||
| Line 45: | Line 46: | ||
*NoVLP was kindly given to us by Dr. Daisuke Sano in Hokkaido University. | *NoVLP was kindly given to us by Dr. Daisuke Sano in Hokkaido University. | ||
*scFv-encoding plasmid (NoV GII.4 strain-specific antibody 12A2 (vector: pSCCA5-E8d)) was kindly given from Dr. Kyoko Moriguchi of the Fujita Health University Department of Virology Parasitology. | *scFv-encoding plasmid (NoV GII.4 strain-specific antibody 12A2 (vector: pSCCA5-E8d)) was kindly given from Dr. Kyoko Moriguchi of the Fujita Health University Department of Virology Parasitology. | ||
| − | ===== | + | =====Aspect ratio of recombinant '' E.coli ''===== |
When comparing SEM pictures, we noticed that <i>E. coli</i> using INPNC-His system seems to be more elongated compared to those with BclA-His system. To confirm this, we determined aspect ratios of all <i>E. coli</i> from the SEM pictures using ImageJ (see [http://2016.igem.org/Team:Kyoto/Experiments Materials and Methods] for more detail). By Welch’s t-test (two-sided), we found that <i>E. coli</i> with INPNC-His system was in fact more elongated than BclA-His system expressing <i>E. coli</i>. (p=0.0015<0.05). | When comparing SEM pictures, we noticed that <i>E. coli</i> using INPNC-His system seems to be more elongated compared to those with BclA-His system. To confirm this, we determined aspect ratios of all <i>E. coli</i> from the SEM pictures using ImageJ (see [http://2016.igem.org/Team:Kyoto/Experiments Materials and Methods] for more detail). By Welch’s t-test (two-sided), we found that <i>E. coli</i> with INPNC-His system was in fact more elongated than BclA-His system expressing <i>E. coli</i>. (p=0.0015<0.05). | ||
| − | ===== | + | =====Growth curve===== |
| − | We have also made a growth curve of the INPNC-His and BclA-His system expressing <i>E. coli</i>. It shows that INPNC-His-scFv expressing <i>E. coli</i> has significant delays in growth compared to its BclA counterpart | + | We have also made a growth curve of the INPNC-His and BclA-His system expressing <i>E. coli</i>. It shows that INPNC-His-scFv expressing <i>E. coli</i> has significant delays in growth compared to its BclA counterpart. |
| − | [[file:T--Kyoto--projectfig13.png|600px|thumb|center|''' | + | [[file:T--Kyoto--projectfig13.png|600px|thumb|center|'''Fig3''': ''' Growth curve ''' <br>Measurements were done for 10 hours, but the graph only shows 4 hours of measurement since the medium saturated within 4 hours.]] |
| − | + | The results from growth curve and elongated shape both suggests that expression of INPNC-His-scFv to the outer membrane caused significant stress and deformation to the host <i>E. coli</i>. | |
<br><br>Please see <big>[https://parts.igem.org/Part:BBa_K1933100 ☆BBa_K1933100]</big>, [https://parts.igem.org/Part:BBa_K1933101 BBa_K1933101], and [https://parts.igem.org/Part:BBa_K1933200 ☆BBa_K1933200] for full characterization of this part. | <br><br>Please see <big>[https://parts.igem.org/Part:BBa_K1933100 ☆BBa_K1933100]</big>, [https://parts.igem.org/Part:BBa_K1933101 BBa_K1933101], and [https://parts.igem.org/Part:BBa_K1933200 ☆BBa_K1933200] for full characterization of this part. | ||
Latest revision as of 15:21, 19 October 2016
INPNC fused to 6xHis tag
INPNC codes for N- and C- terminal domain of Ice Nucleation Protein (INP) from Pseudomonas syringae , fused to 6xHis tag to be easily identified by Western blotting[1]. This is used as an anchoring motif for the cell surface display of passenger proteins. This domain does not have signal peptide for anchoring on the cell surface[2]. Surface displayed passengers retain enzyme activity even when it is linked to INPNC with 6xHis tag.
For more information, please visit [http://2016.igem.org/Team:Kyoto our wiki]
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 952
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 72
Illegal NgoMIV site found at 405
Illegal AgeI site found at 823 - 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Ice Nucleation Protein(INP) used for our surface display systems is produced by P. syringae , a famous model organism among ice nucleation active (INA) bacteria.
Although the freezing point of water is 0°C, pure water can supercool to -40°C before homogeneous ice nucleation randomly occur to form ice crystals. Generally, minimally impure water will freeze spontaneously before -15°C.
However, INP produced by P. syringae causes ice nucleation as high as -2°C. This bacterium uses this property to form frosts on plants, causing damage and making the plants release nutrients for their growth[3].
INP is expressed as the ice nucleation site, expressed in the outer cell membrane. Modified INP is used as a surface expression system.
We omitted stop codon from this part to make BBa_K1933011.
We used BBa_K1933011 for registration of ☆BBa_K1933100, BBa_K1933101, and ☆BBa_K1933200.
Characterization
Sequence confirmed!
We further characterized functionality and improved BBa_K811005 from Penn 2012!!
Western blotting
We added His-tag to BBa_K811005. This addition introduces two new functions to the fusion protein:
First, we can use anti-His tag antibody to detect their surface expression regardless of passenger protein placed downstream. We confirmed this with detections of the INPNC-His-(passenger protein) by Western blotting using anti-His tag antibody(Fig1(a)).
Second, INPNC-His-(passenger protein) can be purified using His tag’s affinity to nickel columns even in denaturing conditions to recollect it from membrane fractions of E.coli . This would allow fusion protein detection even if the protein’s expression levels are low, further strengthening the first function. We tested this function by Western blotting. Sample was prepared by obtaining the membrane fraction from the E. coli lysate by ultracentrifugation, and solubilized them using 7M guanidine-HCl, then purification of the target protein using Nickel Sepharose(Fig1(b)).
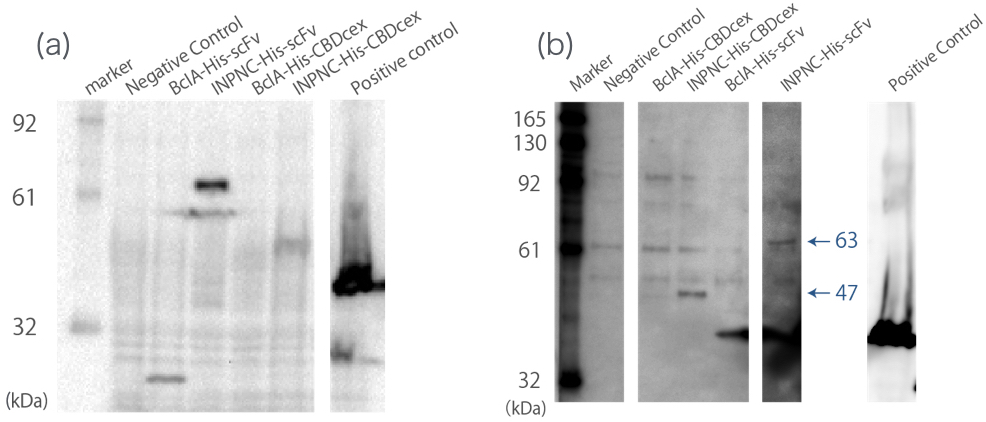
Negative Control:BBa_K165002, BclA-His-scFv:33 kDa, INPNC-His-scFv:63 kDa, BclA-His-CBDcex:17 kDa, INPNC-His-CBDcex:47 kDa, Positive control(Lpp-OmpA-scFv-His):43 kDa
These two results confirms the enhanced functions of INPNC parts (BBa_K811005) existing in the iGEM Regisrtry.
Scanning Electron Microscopy
We constructed INPNC-His-scFv (scFv:a protein made from fusing a variable region of the antibody, VH, and VL, with a linker peptide) and BclA-His-scFv (BclA:another surface display module) encoding plasmids using this part. Then we transformed E. coli (strain:DH5α)with these plasmids, and observed its interaction with Norovirus-like particles (NoVLP, the capsid proteins of NoV) of NoV GII.4 strain. (Fig2) We observed NoVLP only on the surface of INPNC-His-scFv expressing E. coli , as NoVLP could not be observed in E.coli expressing BclA-His-scFv, (which may be due to low expression levels, seen in Fig2), this NoVLP is not unspecifically mixed with the sample. Thus we concluded that we observed the binding of our INPNC-His-scFv expressing E. coli to NoVLP.
Thus, we conclude that this part successfully worked.
- NoVLP was kindly given to us by Dr. Daisuke Sano in Hokkaido University.
- scFv-encoding plasmid (NoV GII.4 strain-specific antibody 12A2 (vector: pSCCA5-E8d)) was kindly given from Dr. Kyoko Moriguchi of the Fujita Health University Department of Virology Parasitology.
Aspect ratio of recombinant E.coli
When comparing SEM pictures, we noticed that E. coli using INPNC-His system seems to be more elongated compared to those with BclA-His system. To confirm this, we determined aspect ratios of all E. coli from the SEM pictures using ImageJ (see [http://2016.igem.org/Team:Kyoto/Experiments Materials and Methods] for more detail). By Welch’s t-test (two-sided), we found that E. coli with INPNC-His system was in fact more elongated than BclA-His system expressing E. coli. (p=0.0015<0.05).
Growth curve
We have also made a growth curve of the INPNC-His and BclA-His system expressing E. coli. It shows that INPNC-His-scFv expressing E. coli has significant delays in growth compared to its BclA counterpart.
The results from growth curve and elongated shape both suggests that expression of INPNC-His-scFv to the outer membrane caused significant stress and deformation to the host E. coli.
Please see ☆BBa_K1933100, BBa_K1933101, and ☆BBa_K1933200 for full characterization of this part.
Judging
We fulfilled criteria listed below with this part.
- Validated Part/ Validated Contribution(Silver)
- Improve a previous part or project(Gold)
- [http://2016.igem.org/Team:Kyoto/Proof Proof of concept](Gold)
Reference
[1]Yim, Sung-Kun, et al. "Surface display of heme-and diflavin-containing cytochrome P450 BM3 in Escherichia coli : a whole cell biocatalyst for oxidation." Journal of microbiology and biotechnology, 20.4, (2010): 712-717.
[2]Park, Tae Jung, et al. "Surface display of recombinant proteins on Escherichia coli by BclA exosporium of Bacillus anthracis. " Microbial cell factories, 12.1, (2013): 1.
[3]Alexander M. McCorkle. "Natural Ice-Nucleating Bacteria Increase the Freezing Tolerance of the Intertidal Bivalve Geukensia demissa." Biology Honors Papers, 1.5,(2009): 1.


