Difference between revisions of "Part:BBa K2065003"
Tjvsonsbeek (Talk | contribs) (→Characterization) |
Tjvsonsbeek (Talk | contribs) |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 14: | Line 14: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K2065003 short</partinfo> | <partinfo>BBa_K2065003 short</partinfo> | ||
| − | + | This BioBrick is a coding sequence for the CT52 protein, attached to the caspase-9 protein with a GGS10 linker in between. Because CT52 has a binding interaction for the T14-3-3 scaffold protein under influence of the small molecule fusicoccin, it can be used for assembly of proteins on this scaffold protein. | |
| − | + | ||
===Design of the construct=== | ===Design of the construct=== | ||
| − | This parts is constructed of three subparts; CT52, a GGS10 linker and | + | This parts is constructed of three subparts; CT52, a GGS10 linker and caspase-9. It has a molecular weight of 52 kDa. CT52 is the name for the isolated last 52 C-terminal amino acids of the plant plasma H+-ATPase PMA2 [1]. The N-terminus of CT52 is connected to a flexible GGS10 linker. This linker consists of a ten times repeating Glycine-Glycine-Serine amino acid sequence. |
===Usage=== | ===Usage=== | ||
| − | Caspase 9 is an apoptosis inducer which can activates by dimerizing with itself. The ability of caspase 9 to cleave the Ac-LEHD-AFC substrate can be used to determine the caspase 9 activity. When two caspase-9 proteins dimerize, and cleave Ac-LEHD-AFC, this substrate splits into a non-fluorescent peptide and a fluorescent part. This fluorescence has an excitation of 400 nm and emission at 505 nm. Thus caspase 9 activity can be measured by means of fluorescence. [1] | + | Caspase-9 is an apoptosis inducer which can activates by dimerizing with itself. The ability of caspase-9 to cleave the Ac-LEHD-AFC substrate can be used to determine the caspase 9 activity. When two caspase-9 proteins dimerize, and cleave Ac-LEHD-AFC, this substrate splits into a non-fluorescent peptide and a fluorescent part. This fluorescence has an excitation of 400 nm and emission at 505 nm. Thus caspase-9 activity can be measured by means of fluorescence. [1] |
| − | When caspase-9 and CT52 are linked, the binding affinity of CT52 and the T14-3-3 construct can be measured by means of | + | When caspase-9 and CT52 are linked, the binding affinity of CT52 and the T14-3-3 construct can be measured by means of fluorescence. Because caspase-9 is an apoptosis inducer, CT52-caspase9 could also be used in a T14-3-3 based kill switch. |
| + | |||
| + | [[File:T--TU-Eindhoven--Caspase9AssayBB.png]] | ||
| + | <br> | ||
| + | ''Figure 1: Caspase-9 activation with CT52-Caspase9, T14-3-3 homotetramer and Ac-LEHD-AFC'' | ||
| + | |||
===Sequence=== | ===Sequence=== | ||
| − | The sequence of our | + | The sequence of our CT52-Capase9 has been verified by StarSeq. It contains the prefix and suffix with the correct restriction sites (EcoRI, XbaI, SpeI and PstI). CT52-caspase9 is 1386 bp long. The specifications of the protein can be found below. |
[[File:T--TU-Eindhoven--Caspase9Snap.png]] | [[File:T--TU-Eindhoven--Caspase9Snap.png]] | ||
| − | + | <br> | |
| + | ''Figure 2: Snapgene map of BBa_K2065003'' | ||
| Line 36: | Line 41: | ||
===Characterization=== | ===Characterization=== | ||
| − | iGEM TU Eindhoven planned to use this part to characterize a tetrameric variant of the T14-3-3 scaffold protein. The constructing of this protein succeeded on DNA level, but due to time limitations the protein couldn't be expressed. To show the caspase 9 dimerization this was tested on a homodimeric T14-3-3 scaffold protein. | + | iGEM TU Eindhoven planned to use this part to characterize a tetrameric variant of the T14-3-3 scaffold protein. The constructing of this protein succeeded on DNA level, but due to time limitations the protein couldn't be expressed. To show the caspase-9 dimerization this was tested on a homodimeric T14-3-3 scaffold protein. This shows caspase-9 activation on T14-3-3 scaffolds is possible. |
| + | |||
| + | |||
| + | [[File:T--TU-Eindhoven--Caspase9BB.png]] | ||
| + | <br> | ||
| + | ''Figure 3: T14-3-3 homodimer caspase-9 assay. The concentration of scaffold is varied from 0 nM to 1 µM. The concentration of CT52-caspase-9 used is 200 nM and the concentration of fusicoccin used is 50 µM. On the x-axis the slope of the measured fluorescence is presented.'' | ||
| + | <br> | ||
===Protein Specifications=== | ===Protein Specifications=== | ||
Latest revision as of 09:28, 19 October 2016
CT52-Caspase9 This BioBrick is a coding sequence for the CT52 protein, attached to the caspase-9 protein with a GGS10 linker in between. Because CT52 has a binding interaction for the T14-3-3 scaffold protein under influence of the small molecule fusicoccin, it can be used for assembly of proteins on this scaffold protein.
Design of the construct
This parts is constructed of three subparts; CT52, a GGS10 linker and caspase-9. It has a molecular weight of 52 kDa. CT52 is the name for the isolated last 52 C-terminal amino acids of the plant plasma H+-ATPase PMA2 [1]. The N-terminus of CT52 is connected to a flexible GGS10 linker. This linker consists of a ten times repeating Glycine-Glycine-Serine amino acid sequence.
Usage
Caspase-9 is an apoptosis inducer which can activates by dimerizing with itself. The ability of caspase-9 to cleave the Ac-LEHD-AFC substrate can be used to determine the caspase 9 activity. When two caspase-9 proteins dimerize, and cleave Ac-LEHD-AFC, this substrate splits into a non-fluorescent peptide and a fluorescent part. This fluorescence has an excitation of 400 nm and emission at 505 nm. Thus caspase-9 activity can be measured by means of fluorescence. [1] When caspase-9 and CT52 are linked, the binding affinity of CT52 and the T14-3-3 construct can be measured by means of fluorescence. Because caspase-9 is an apoptosis inducer, CT52-caspase9 could also be used in a T14-3-3 based kill switch.
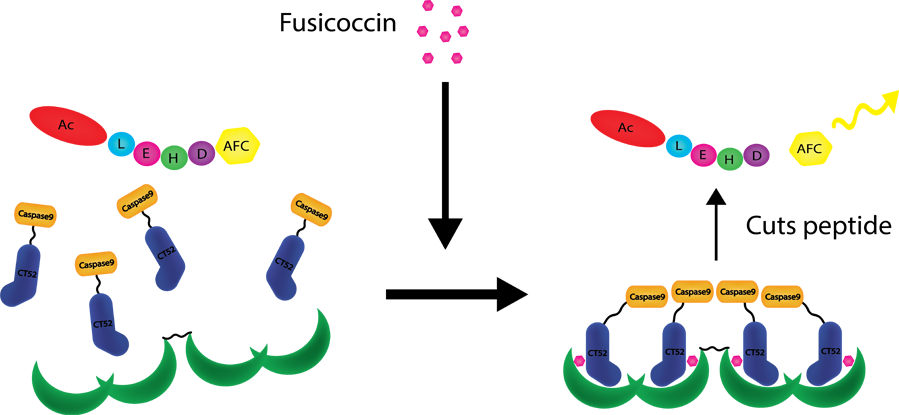
Figure 1: Caspase-9 activation with CT52-Caspase9, T14-3-3 homotetramer and Ac-LEHD-AFC
Sequence
The sequence of our CT52-Capase9 has been verified by StarSeq. It contains the prefix and suffix with the correct restriction sites (EcoRI, XbaI, SpeI and PstI). CT52-caspase9 is 1386 bp long. The specifications of the protein can be found below.
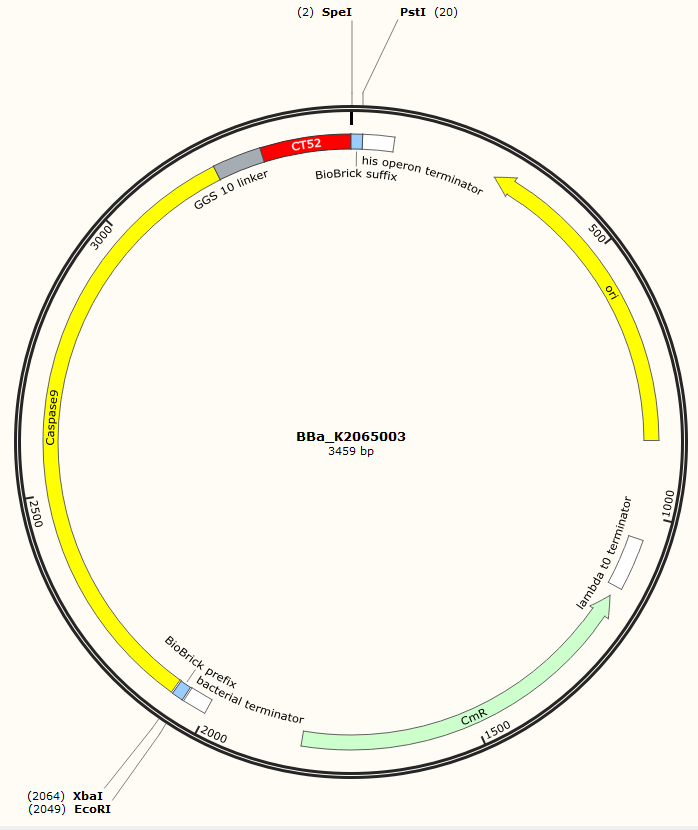
Figure 2: Snapgene map of BBa_K2065003
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
iGEM TU Eindhoven planned to use this part to characterize a tetrameric variant of the T14-3-3 scaffold protein. The constructing of this protein succeeded on DNA level, but due to time limitations the protein couldn't be expressed. To show the caspase-9 dimerization this was tested on a homodimeric T14-3-3 scaffold protein. This shows caspase-9 activation on T14-3-3 scaffolds is possible.
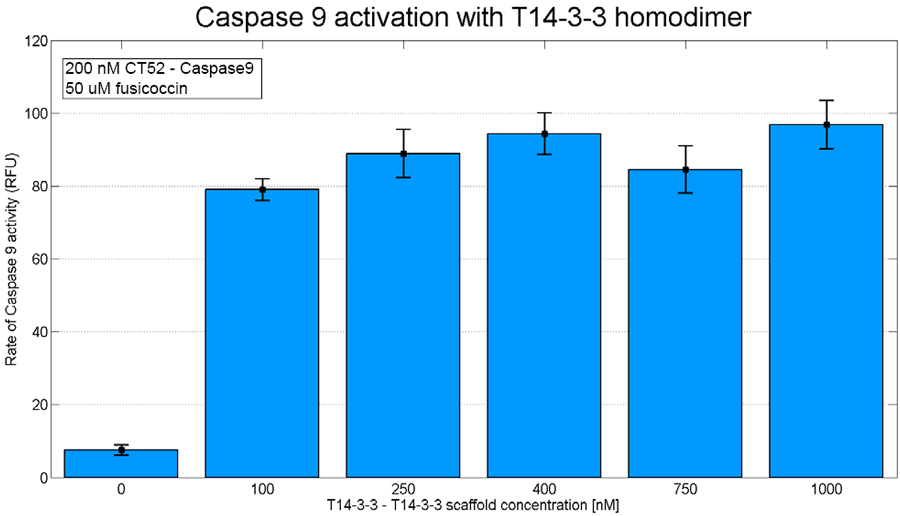
Figure 3: T14-3-3 homodimer caspase-9 assay. The concentration of scaffold is varied from 0 nM to 1 µM. The concentration of CT52-caspase-9 used is 200 nM and the concentration of fusicoccin used is 50 µM. On the x-axis the slope of the measured fluorescence is presented.
Protein Specifications
| Protein Specifications | |||
|---|---|---|---|
| General Information | Number of amino acids | 475 | |
| Molecular weight | 51624 | ||
| Theoretical pi | 5.01 | ||
| Extinction coefficient | 27555 | ||
| Formula | C2248H3538N626O730S19 | ||
| Total numbers of atons | 7161 | ||
| Amino Acid Composition | Amino Acid | Frequency | Percentage(%) |
| Ala(A) | 26 | 5.5 | |
| Arg(R | 12 | 4.4 | |
| Asn(N) | 16 | 3.4 | |
| Asp(D) | 27 | 5.7 | |
| Cys(C) | 11 | 2.3 | |
| Gln(Q) | 20 | 4.2 | |
| Glu(E) | 43 | 9.1 | |
| Gly(G) | 52 | 10.9 | |
| His(H) | 11 | 2.3 | |
| Ile(I) | 24 | 5.1 | |
| Leu(L) | 40 | 8.4 | |
| Lys(K) | 27 | 5.7 | |
| Met(M) | 8 | 1.7 | |
| Phe(F) | 23 | 4.8 | |
| Pro(P) | 20 | 4.2 | |
| Ser(S) | 44 | 9.3 | |
| Thr(T) | 27 | 5.7 | |
| Trp(W) | 3 | 0.6 | |
| Tyr(T) | 7 | 1.5 | |
| Val(V) | 25 | 5.3 | |
| Pyl(O) | 0 | 0.0 | |
| Sec(U) | 0 | 0.0 | |
References
[1] Pop, C., Timmer, J., Sperandio, S., & Salvesen, G. (2006). The Apoptosome Activates Caspase-9 by Dimerization. Molecular Cell, 22(2), 269-275.
