Difference between revisions of "Part:BBa K2065005"
Tjvsonsbeek (Talk | contribs) (→Verification of the scaffold) |
|||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 15: | Line 15: | ||
<partinfo>BBa_K2065005 short</partinfo> | <partinfo>BBa_K2065005 short</partinfo> | ||
| − | The T14-3-3 – T14-3-3(E19R) | + | The T14-3-3 – T14-3-3(E19R) consists of a wildtype monomer and a monomer with mutation E19R, where the 19th amino acid is changed from glutamic acid to arginine. This heterodimer is a scaffold protein and originates from the Tobacco plant. Protein-Protein Interactions (PPI) can be induced on this scaffold by linking proteins of choice to the CT52 protein, a protein with a known interaction with this scaffold protein. |
| Line 54: | Line 54: | ||
The sequence of our heterodimer T14-3-3 has been verified by StarSeq. It contains the prefix and suffix with the correct restriction sites (EcoRI, XbaI, SpeI and PstI). Unfortunately this BioBrick contains 2 extra EcoRI restriction sites, it is important to be aware of this if you plan to use it. T14-3-3 (E19R)-T14-3-3 is 1577 bp long. | The sequence of our heterodimer T14-3-3 has been verified by StarSeq. It contains the prefix and suffix with the correct restriction sites (EcoRI, XbaI, SpeI and PstI). Unfortunately this BioBrick contains 2 extra EcoRI restriction sites, it is important to be aware of this if you plan to use it. T14-3-3 (E19R)-T14-3-3 is 1577 bp long. | ||
[[File:T--TU-Eindhoven--E19RSnap.png]] | [[File:T--TU-Eindhoven--E19RSnap.png]] | ||
| − | + | <br> | |
| − | + | ''Figure 1: Snapgene map of BBa_K2065005.'' | |
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
| Line 66: | Line 66: | ||
===Verification of the scaffold=== | ===Verification of the scaffold=== | ||
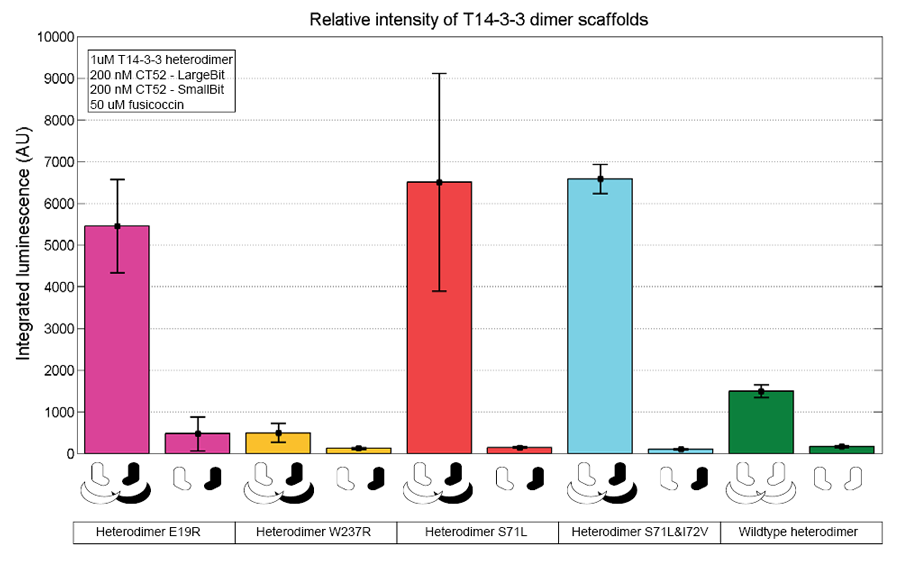
| − | To verify whether the scaffold shows the expected activity the activity of the scaffold at different concentrations was measured. Below a bar graph with the integrated | + | To verify whether the scaffold shows the expected activity the activity of the scaffold at different concentrations was measured. This confirms that T14-3-3 - T14-3-3(E19R) is a functional scaffold. |
| + | |||
| + | [[File:T--TU-Eindhoven--RelinsCom.png]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | |||
| + | ''Figure 2: T14-3-3 functionality measurement representing the relative intensities from each mutation set. The scaffold concentrations used are 1 µM. 50 µM fusicoccin was used for all measurements. CT52-LargeBiT and CT52-SmallBiT were used with a concentration of 200 nM, CT52-LargeBiT contained the mutation. The substrate used was 1250x diluted from stock. '' | ||
| + | |||
| + | To verify whether the scaffold shows the expected activity the activity of the scaffold at different concentrations was measured. Below a bar graph with the integrated bioluminescencescence over a timespan of 20 minutes. This reinforces that T14-3-3 - T14-3-3(E19R) is a functional scaffold. | ||
| + | |||
| + | |||
| + | |||
| + | [[File:T--TU-Eindhoven--ERgrad.png]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | ''Figure 3: T14-3-3 functionality measurement with varying scaffold concentrations from 0 nM to 1 µM T14-3-3(E19R)-T14-3-3. 50 µM fusicoccin was used for all measurements. Mutant forms CT52 (K943D)-LargeBiT and CT52(wildtype)-SmallBiT were used with a concentration of 200 nM. The substrate used was 1250x diluted from stock.'' | ||
===Orthogonality of the scaffold=== | ===Orthogonality of the scaffold=== | ||
| − | To check whether this heterodimeric scaffold protein is orthogonal, the following was done. The magnitude of the integrated luminescence for complementary CT52-split NanoLuc proteins ( CT52(K943D)-LargeBiT and CT52-SmallBit) was compared with three other situations. The first one consisted of two mutated CT52-split NanoLuc proteins (CT52(K943D)-LargeBiT and CT52(K943D)-SmallBit) instead of one wildtype and one mutated CT52-split nanoluc protein. The second one consisted of two wildtype CT52-split NanoLuc proteins (CT52-LargeBiT and CT52-SmallBit). The last one consisted of the two complementary CT52-split NanoLuc proteins without scaffold present. As a negative control substrate without scaffold protein of CT52-split NanoLuc proteins was used. | + | To check whether this heterodimeric scaffold protein is orthogonal, the following was done. The magnitude of the integrated luminescence for complementary CT52-split NanoLuc proteins ( CT52(K943D)-LargeBiT and CT52-SmallBit) was compared with three other situations. The first one consisted of two mutated CT52-split NanoLuc proteins (CT52(K943D)-LargeBiT and CT52(K943D)-SmallBit) instead of one wildtype and one mutated CT52-split nanoluc protein. The second one consisted of two wildtype CT52-split NanoLuc proteins (CT52-LargeBiT and CT52-SmallBit). The last one consisted of the two complementary CT52-split NanoLuc proteins without scaffold present. As a negative control substrate without scaffold protein of CT52-split NanoLuc proteins was used. |
| + | [[File:T--TU-Eindhoven--SLIVortho.png]] | ||
| + | <br> | ||
| + | ''Figure 4: T14-3-3 orthogonality measurement with 1 µM T14-3-3(W237R)-T14-3-3 concentration for bar 1-3, 0 µM T14-3-3(W237R)-T14-3-3 for bar 4 and 5. 50 µM fusicoccin for bar 1 -5. Mutant forms CT52-LargeBiT and CT52-SmallBiT are present in different combinations. Bar 1: CT52(K943D)-LargeBiT,CT52(wildtype)-SmallBiT. Bar 2: CT52(K943D)-LargeBiT,CT52(K943D)-SmallBiT. Bar 3: CT52(wildtype)-LargeBiT,CT52(wildtype)-SmallBiT. Bar 4: CT52(K943D)-LargeBiT,CT52-SmallBiT. Bar 5: No CT52-LargeBiT or CT52-SmallBiT present. The substrate used was 1250x diluted from stock. '' | ||
===Protein Specifications=== | ===Protein Specifications=== | ||
Latest revision as of 00:46, 19 October 2016
T14-3-3 - T14-3-3 (E19R)
The T14-3-3 – T14-3-3(E19R) consists of a wildtype monomer and a monomer with mutation E19R, where the 19th amino acid is changed from glutamic acid to arginine. This heterodimer is a scaffold protein and originates from the Tobacco plant. Protein-Protein Interactions (PPI) can be induced on this scaffold by linking proteins of choice to the CT52 protein, a protein with a known interaction with this scaffold protein.
Biology of the construct
The 14-3-3 proteins are a family of proteins which are well preserved in evolution and are present in all eukaryotic cells. From the fact that there is a high diversity of interaction partners of the 14-3-3 protein, can be concluded that 14-3-3 contributes in many cell processes.[1] Examples of regulating and coordinating cell processes where 14-3-3 plays a role are cell cycle progression, apoptosis, metabolism, transcription, regulation of gene expression and DNA damage repair.
A variant of the 14-3-3 protein originates from the Nicotiana plumbaginifolia (Tobacco) plant and is called T14-3-3. [1] T14-3-3 proteins consist of two identical monomers, that dimerize to form a functional scaffold. Each monomer contains a bundle of nine antiparallel alfa-helices. Helices H3, H5, H7 and H9 form the “amphipathic ligand-binding groove”, in which other proteins can bind and interact with their co-protein. [2]
A protein that shows a high affinity for binding in the groove of T14-3-3 is called CT52. This protein consist of the last 52 amino acids of the C-terminal region of the H+-ATPase. The N-terminus of CT52 is a free end, which can be used to link proteins to. The binding between CT52 and T14-3-3 is stabilized by the small molecule Fusicoccin. When Fusicoccin is present, the affinity of CT52 for T14-3-3 is increased in 30 fold with respect to the situation when Fusicoccin is not present [3]. The binding interaction with Fusicoccin is reversibel, this means CT52 will dissociate from the scaffold.
Gene Design and Usage
To construct a T14-3-3 dimer, two monomers are linked to each other with a flexible GGS10 linker. This linker consists of a ten times repeating glycine-glycine-serine amino acid sequence.
To construct a T14-3-3 dimer, two monomers are linked to each other with a flexible GGS10 linker. This linker consists of a ten times repeating glycine-glycine-serine amino acid sequence.
A homodimeric T14-3-3 scaffold consists of two identical monomers, which enables the assembly of two identical CT52-protein complexes. To enable the assembly of two different CT52-protein complexes, this heterodimeric variant of T14-3-3 is created. To achieve this, a mutation is introduced in one monomer of the dimer. This mutation has to be orthogonal with respect to the CT52 protein, to ensure minimal interaction with the natural processes in that organism.
One of the previous mutations that was found for T14-3-3 by Skwarczynska et al. (2013) is E19R [4]. This means that the 19th amino acid is changed from the negatively charged glutamic acid to positively charged arginine. To ensure orthogonality a corresponding mutation in CT52 was found, K943D. Where positively charged lysine is changed for negatively charged aspartic acid. So a complementary charge exchange is conducted.
The T14-3-3 dimer consists of a wildtype monomer and a monomer that contains the mutation E19R. The scaffold protein can now bind two different CT52-protein complexes.
In the project of iGEM TU Eindhoven, T14-3-3 – T14-3-3 (E19R) was used as an positive control to test other binding modifications that were introduced to the T14-3-3 scaffold, these parts are also documented as BioBricks namely, BBa_K2065002 , <a href="">BBa_K2065004</a> and <a href="">BBa_K2065006</a>. The specifications of the T14-3-3 – T14-3-3(E19R) protein can be found below.
Sequence
The sequence of our heterodimer T14-3-3 has been verified by StarSeq. It contains the prefix and suffix with the correct restriction sites (EcoRI, XbaI, SpeI and PstI). Unfortunately this BioBrick contains 2 extra EcoRI restriction sites, it is important to be aware of this if you plan to use it. T14-3-3 (E19R)-T14-3-3 is 1577 bp long.

Figure 1: Snapgene map of BBa_K2065005.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 1234
Illegal EcoRI site found at 1389 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 1234
Illegal EcoRI site found at 1389 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 1234
Illegal EcoRI site found at 1389 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 1234
Illegal EcoRI site found at 1389 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 1234
Illegal EcoRI site found at 1389 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 937
Characterization
To verify the orthogonality and activity of T14-3-3 – T14-3-3(E19R) scaffold, in vitro assays have been performed with NanoLuc split luciferases linked to CT52. The NanoLuc reporter system consists of a small and a large bit, when those two are dimerized they are able to activate an luminogenic substrate which emits luminescence at a wavelength of 460 nm. The small and large bit of this NanoLuc reporter system are called: SmallBiT and LargeBiT. The BioBricks of this NanoLuc reporter system linked to CT52 are <a href="">BBa_K2065000</a> and <a href="">BBa_K2065007</a>.
Verification of the scaffold
To verify whether the scaffold shows the expected activity the activity of the scaffold at different concentrations was measured. This confirms that T14-3-3 - T14-3-3(E19R) is a functional scaffold.
Figure 2: T14-3-3 functionality measurement representing the relative intensities from each mutation set. The scaffold concentrations used are 1 µM. 50 µM fusicoccin was used for all measurements. CT52-LargeBiT and CT52-SmallBiT were used with a concentration of 200 nM, CT52-LargeBiT contained the mutation. The substrate used was 1250x diluted from stock.
To verify whether the scaffold shows the expected activity the activity of the scaffold at different concentrations was measured. Below a bar graph with the integrated bioluminescencescence over a timespan of 20 minutes. This reinforces that T14-3-3 - T14-3-3(E19R) is a functional scaffold.
Figure 3: T14-3-3 functionality measurement with varying scaffold concentrations from 0 nM to 1 µM T14-3-3(E19R)-T14-3-3. 50 µM fusicoccin was used for all measurements. Mutant forms CT52 (K943D)-LargeBiT and CT52(wildtype)-SmallBiT were used with a concentration of 200 nM. The substrate used was 1250x diluted from stock.
Orthogonality of the scaffold
To check whether this heterodimeric scaffold protein is orthogonal, the following was done. The magnitude of the integrated luminescence for complementary CT52-split NanoLuc proteins ( CT52(K943D)-LargeBiT and CT52-SmallBit) was compared with three other situations. The first one consisted of two mutated CT52-split NanoLuc proteins (CT52(K943D)-LargeBiT and CT52(K943D)-SmallBit) instead of one wildtype and one mutated CT52-split nanoluc protein. The second one consisted of two wildtype CT52-split NanoLuc proteins (CT52-LargeBiT and CT52-SmallBit). The last one consisted of the two complementary CT52-split NanoLuc proteins without scaffold present. As a negative control substrate without scaffold protein of CT52-split NanoLuc proteins was used.
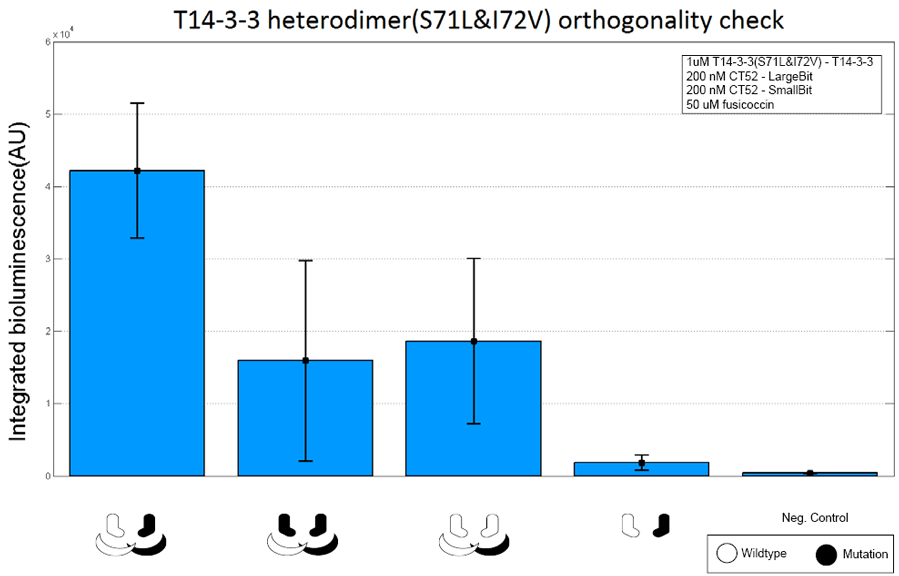
Figure 4: T14-3-3 orthogonality measurement with 1 µM T14-3-3(W237R)-T14-3-3 concentration for bar 1-3, 0 µM T14-3-3(W237R)-T14-3-3 for bar 4 and 5. 50 µM fusicoccin for bar 1 -5. Mutant forms CT52-LargeBiT and CT52-SmallBiT are present in different combinations. Bar 1: CT52(K943D)-LargeBiT,CT52(wildtype)-SmallBiT. Bar 2: CT52(K943D)-LargeBiT,CT52(K943D)-SmallBiT. Bar 3: CT52(wildtype)-LargeBiT,CT52(wildtype)-SmallBiT. Bar 4: CT52(K943D)-LargeBiT,CT52-SmallBiT. Bar 5: No CT52-LargeBiT or CT52-SmallBiT present. The substrate used was 1250x diluted from stock.
Protein Specifications
| Protein Specifications | |||
|---|---|---|---|
| General Information | Number of amino acids | 514 | |
| Molecular weight | 56798.79 | ||
| Theoretical pi | 5.02 | ||
| Extinction coefficient | 55030 of 54780 | ||
| Formula | C2478H3949N983O807S18 | ||
| Total numbers of atons | 7935 | ||
| Amino Acid Composition | Amino Acid | Frequency | Percentage(%) |
| Ala(A) | 53 | 10.3 | |
| Arg(R | 29 | 5.6 | |
| Asn(N) | 22 | 4.3 | |
| Asp(D) | 22 | 4.3 | |
| Cys(C) | 4 | 0.8 | |
| Gln(Q) | 12 | 2.3 | |
| Glu(E) | 59 | 11.5 | |
| Gly(G) | 39 | 7.6 | |
| His(H) | 6 | 1.2 | |
| Ile(I) | 28 | 5.4 | |
| Leu(L) | 52 | 10.1 | |
| Lys(K) | 32 | 6.2 | |
| Met(M) | 14 | 2.7 | |
| Phe(F) | 12 | 2.3 | |
| Pro(P) | 10 | 1.9 | |
| Ser(S) | 50 | 9.7 | |
| Thr(T) | 24 | 4.7 | |
| Trp(W) | 4 | 0.8 | |
| Tyr(T) | 22 | 4.3 | |
| Val(V) | 20 | 3.9 | |
| Pyl(O) | 0 | 0.0 | |
| Sec(U) | 0 | 0.0 | |
References
[1] – Ottmann, C., Marco, S., Jaspert, N., Marcon, C., Schauer, N., Weyand, M., Vandermeeren, C., Duby, G., Boutry, M., Wittinghofer, A., Rigaud, J. and Oecking, C. (2007). Structure of a 14-3-3 Coordinated Hexamer of the Plant Plasma Membrane H+-ATPase by Combining X-Ray Crystallography and Electron Cryomicroscopy. Molecular Cell, 25(3), pp.427-440.
[2] - Obsil, T. and Obsilova, V. (2011). Structural basis of 14-3-3 protein functions. Seminars in Cell & Developmental Biology, 22(7), pp.663-672.
[3] - Milroy, L., Brunsveld, L., & Ottmann, C. (2013). Stabilization and Inhibition of Protein–Protein Interactions: The 14-3-3 Case Study. ACS Chem. Biol., 8(1), 27-35.
[4] - Skwarczynska, M., Molzan, M., & Ottmann, C. (2013). Activation of NF-κB signalling by Fusicoccin-induced dimerization.. Proc Natl Acad Sci U S A., 110(5), 377-386. doi:10.1073/pnas.1212990110