Difference between revisions of "Part:BBa K1998009"
SWinchester (Talk | contribs) (→Protein information) |
Ari edmonds (Talk | contribs) |
||
| (14 intermediate revisions by 2 users not shown) | |||
| Line 13: | Line 13: | ||
===Overview=== | ===Overview=== | ||
| − | |||
HYD1 is an oxygen-tolerant hydrogenase; it is a respiratory enzyme that catalyses hydrogen oxidation. It has been suggested that it functions at more positive redox potentials, which are located at the aerobic-anaerobic interface. It forms one of the enzymes found in our hydrogenase pathway of our system. | HYD1 is an oxygen-tolerant hydrogenase; it is a respiratory enzyme that catalyses hydrogen oxidation. It has been suggested that it functions at more positive redox potentials, which are located at the aerobic-anaerobic interface. It forms one of the enzymes found in our hydrogenase pathway of our system. | ||
<br><br> | <br><br> | ||
| − | + | <html><center><img src="https://static.igem.org/mediawiki/parts/b/b5/HydrogenPathwayUpdated2016.jpeg" alt="HydrogenProduction" height="50%"width="75%"></center></html> | |
===Biology & Literature=== | ===Biology & Literature=== | ||
| − | + | More specifically <i>Hyd1</i> catalyses the oxidation of hydrogen in the anaerobic respiration of <i>E.coli</i> [2] this done by working in parallel with <i>Hyd</i> 2 before fermentation is undertaken by <i>Hyd3</i> [2, 3]. <i>Hyd1</i> is a membrane bound hydrogenase that facilitates the uptake of oxidated hydrogen gas [3]. The <i>hya</i> operon encodes the Hydrogenase peptide which is initiated under anaerobic conditions as well as acidic pH levels which signals that H+ must be facilitated to move from ICF to ECF and viceverser to maintain pH levels in <i>E.coli</i> [3, 4]. | |
| − | < | + | |
| − | < | + | |
| − | + | ||
<br> | <br> | ||
| + | |||
| + | ===Part Verification=== | ||
| + | |||
| + | <html><center><img src="https://static.igem.org/mediawiki/2016/b/bd/T--Macquarie_Australia--HYDShowGel.png" " width="50%" height="35%"></center> </html> | ||
| + | <b>Fig 1.</b> A gel consisting of EcorI/PstI double digests for the hydEF part (Lane 4) The hydEF (3611 bp) was observed as expected in the gel. | ||
===Protein information=== | ===Protein information=== | ||
| − | + | <i>Hyd1</i><br> | |
| − | + | Mass: 53.13 kDa<br> | |
| − | + | Sequence: <br> | |
| − | + | MSALVLKPCAAVSIRGSSCRARQVAPRAPLAASTVRVALATLEAPARRLGNVACAAAAPAAEAPLSHVQQALAELAKPKDDPTRKHVCVQVAPAVRVAIAETLGLAPGATT | |
| − | + | PKQLAEGLRRLGFDEVFDTLFGADLTIMEEGSELLHRLTEHLEAHPHSDEPLPMFTSCCPGWIAMLEKSYPDLIPYVSSCKSPQMMLAAMVKSYLAEKKGIAPKDMVMV | |
| − | <br> | + | SIMPCTRKQSEADRDWFCVDADPTLRQLDHVITTVELGNIFKERGINLAELPEGEWDNPMGVGSGAGVLFGTTGGVMEAALRTAYELFTGTPLPRLSLSEVRGMDGIKET |
| − | <br> | + | NITMVPAPGSKFEELLKHRAAARAEAAAHGTPGPLAWDGGAGFTSEDGRGGITLRVAVANGLGNAKKLITKMQAGEAKYDFVEIMACPAGCVGGGGQPRSTDKAITQKR |
| + | QAALYNLDEKSTLRRSHENPSIRELYDTYLGEPLGHKAHELLHTHYVAGGVEEKDEKK | ||
<br> | <br> | ||
===References=== | ===References=== | ||
| − | + | [1] Nair RR, Emmons MF, Cress AE, Argilagos RF, Lam K, Kerr WT, Wang HG, Dalton WS, Hazlehurst LA. HYD1-induced increase in reactive oxygen species leads to autophagy and necrotic cell death in multiple myeloma cells. Molecular cancer therapeutics. 2009 Aug 1;8(8):2441-51. | |
| − | [1] | + | <br><br> |
| − | <br> | + | [2] Redwood MD, Mikheenko IP, Sargent F, Macaskie LE. Dissecting the roles of Escherichia coli hydrogenases in biohydrogen production. FEMS microbiology letters. 2008 Jan 1;278(1):48-55. |
| − | [2] | + | <br><br> |
| − | <br> | + | [3] Bisaillon A, Turcot J, Hallenbeck PC. The effect of nutrient limitation on hydrogen production by batch cultures of Escherichia coli. International Journal of Hydrogen Energy. 2006 Sep 30;31(11):1504-8. |
| − | [3] | + | <br><br> |
| − | <br> | + | [4] Trchounian K, Trchounian A. Hydrogenase 2 is most and hydrogenase 1 is less responsible for H 2 production by Escherichia coli under glycerol fermentation at neutral and slightly alkaline pH. international journal of hydrogen energy. 2009 Nov 30;34(21):8839-45. |
| − | [4] | + | |
| − | + | ||
Latest revision as of 01:24, 9 October 2017
Hyd1
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 157
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Overview
HYD1 is an oxygen-tolerant hydrogenase; it is a respiratory enzyme that catalyses hydrogen oxidation. It has been suggested that it functions at more positive redox potentials, which are located at the aerobic-anaerobic interface. It forms one of the enzymes found in our hydrogenase pathway of our system.

Biology & Literature
More specifically Hyd1 catalyses the oxidation of hydrogen in the anaerobic respiration of E.coli [2] this done by working in parallel with Hyd 2 before fermentation is undertaken by Hyd3 [2, 3]. Hyd1 is a membrane bound hydrogenase that facilitates the uptake of oxidated hydrogen gas [3]. The hya operon encodes the Hydrogenase peptide which is initiated under anaerobic conditions as well as acidic pH levels which signals that H+ must be facilitated to move from ICF to ECF and viceverser to maintain pH levels in E.coli [3, 4].
Part Verification
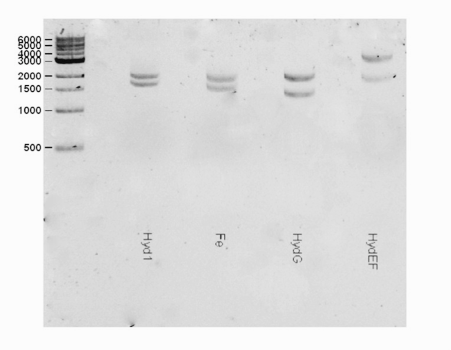
Protein information
Hyd1
Mass: 53.13 kDa
Sequence:
MSALVLKPCAAVSIRGSSCRARQVAPRAPLAASTVRVALATLEAPARRLGNVACAAAAPAAEAPLSHVQQALAELAKPKDDPTRKHVCVQVAPAVRVAIAETLGLAPGATT
PKQLAEGLRRLGFDEVFDTLFGADLTIMEEGSELLHRLTEHLEAHPHSDEPLPMFTSCCPGWIAMLEKSYPDLIPYVSSCKSPQMMLAAMVKSYLAEKKGIAPKDMVMV
SIMPCTRKQSEADRDWFCVDADPTLRQLDHVITTVELGNIFKERGINLAELPEGEWDNPMGVGSGAGVLFGTTGGVMEAALRTAYELFTGTPLPRLSLSEVRGMDGIKET
NITMVPAPGSKFEELLKHRAAARAEAAAHGTPGPLAWDGGAGFTSEDGRGGITLRVAVANGLGNAKKLITKMQAGEAKYDFVEIMACPAGCVGGGGQPRSTDKAITQKR
QAALYNLDEKSTLRRSHENPSIRELYDTYLGEPLGHKAHELLHTHYVAGGVEEKDEKK
References
[1] Nair RR, Emmons MF, Cress AE, Argilagos RF, Lam K, Kerr WT, Wang HG, Dalton WS, Hazlehurst LA. HYD1-induced increase in reactive oxygen species leads to autophagy and necrotic cell death in multiple myeloma cells. Molecular cancer therapeutics. 2009 Aug 1;8(8):2441-51.
[2] Redwood MD, Mikheenko IP, Sargent F, Macaskie LE. Dissecting the roles of Escherichia coli hydrogenases in biohydrogen production. FEMS microbiology letters. 2008 Jan 1;278(1):48-55.
[3] Bisaillon A, Turcot J, Hallenbeck PC. The effect of nutrient limitation on hydrogen production by batch cultures of Escherichia coli. International Journal of Hydrogen Energy. 2006 Sep 30;31(11):1504-8.
[4] Trchounian K, Trchounian A. Hydrogenase 2 is most and hydrogenase 1 is less responsible for H 2 production by Escherichia coli under glycerol fermentation at neutral and slightly alkaline pH. international journal of hydrogen energy. 2009 Nov 30;34(21):8839-45.
