Difference between revisions of "Part:BBa I712009"
(→Results of signal peptide functional analysis) |
|||
| (10 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
| − | Additional characterization of BBaI712009 by [http://2016.igem.org/Team:LMU-TUM_Munich Munich 2016]: | + | =Additional characterization of BBaI712009 by [http://2016.igem.org/Team:LMU-TUM_Munich Munich 2016]:= |
| + | |||
| + | [[File:Muc16_CD4_signal_peptide_001.png|thumb|center|920px| Sequencing results of BioBrick [https://parts.igem.org/wiki/index.php?title=Part:BBa_I712009 BBa_I712009] & consequential ligation problems]] | ||
This part was submitted before the RFC[25] standard was established. Thus this part carries a modified RFC[10] pre- and suffix that changes the reading frame of the suffix in a way that, if it is ligated to a second part, it does not generate a stop codon. Although this approach makes it possible to assemble fusion proteins, the ligation to a second part, which does not carry the modified RFC[10], leads to a frameshift in the second part in a way that it is not functional anymore. | This part was submitted before the RFC[25] standard was established. Thus this part carries a modified RFC[10] pre- and suffix that changes the reading frame of the suffix in a way that, if it is ligated to a second part, it does not generate a stop codon. Although this approach makes it possible to assemble fusion proteins, the ligation to a second part, which does not carry the modified RFC[10], leads to a frameshift in the second part in a way that it is not functional anymore. | ||
| + | Because signal peptides are such important BioBricks we designed new signal peptide parts such as [https://parts.igem.org/wiki/index.php?title=Part:BBa_K2170215 BBa_K2170215] and [https://parts.igem.org/wiki/index.php?title=Part:BBa_K2170214 BBa_K2170214] to help other iGEM teams assembling fusion proteins. The secretion efficiency of the BioBricks was tested by Nano-Glo assay. | ||
| + | ==Results of signal peptide functional analysis== | ||
| + | |||
| + | ===Bioinformatic signal peptide analysis via the SignalP server=== | ||
| + | |||
| + | |||
| + | The [http://www.cbs.dtu.dk/services/SignalP SignalP] server, being able to discriminate between the hydrophobic signal peptide sequence and the hydrophobic transmembrane domain<ref>Petersen, T. N., Brunak, S., von Heijne, G., & Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature methods, 8(10), 785-786.</ref>, can determine whether a sequence possesses the biophysical requirements for functioning as a signal peptide. Furthermore, it is able to predict the probable cleavage site of the signal peptide after its translocation into the ER. For all constructs, signal peptide functionality is predicted, as well as a potential cleavage site. The complete translated amino acid sequences of the respective receptor constructs were used as input. The algorithm for eukaryotes with default D-cutoff value was chosen. | ||
| + | |||
| + | [[File:Muc16 SignalPeptideprediction_002.png|thumb|center|850px| | ||
| + | |||
| + | '''Figure 2''': A) Schematic depiction of a protein-coding mRNA containing a 5'- and 3'-UTR, the first of which being an important factor in translation efficiency. B) Identification of signal peptide functionality and cleavage site within the receptor construct containing the EGFR, Igκ and BM40 signal peptide via the SignalP 4.1 server. Plotted probabilities represent the raw cleavage site “C-score”, the signal peptide “S-score” and the combined cleavage site “Y-score”. C) ClustalW multiple sequence alignment of the BM40, EGFR and Igκ signal peptide construct, depicting a gap for the EGFR-signal peptide construct compared to the newly designed ones, indicating smaller length of the 5'-UTR . For the Igκ and BM40 signal peptide constructs, a T7 promoter spacer as well as a Kozak sequence increase the distance between TATA-box and protein-coding region.]] | ||
| + | |||
| + | For all three signal peptides, the high S-score indicates high signal peptide functionality. For the Igκ signal peptide, a cleavage site between amino acid 20 and 21 within the signal peptide is predicted. For the BM40 signal peptide, a cleavage site between amino acid 17 and 18 is predicted, and for the EGFR signal peptide, a cleavage site between amino acid 24 and 25 is predicted. | ||
| + | </div> | ||
| + | |||
| + | ===Quantification of signal-peptide mediated translocation via a secretion luciferase assay=== | ||
| + | |||
| + | <div class="white-box"> | ||
| + | For the quantification of signal peptide functionality via a luciferase assay, three constructs were created and tested - each containing one of three signal peptides (the EGFR signal peptide, the Igκ signal peptide or the BM40 signal peptide) and a nanoluciferase as well as a CMV promoter, a ''Strep''-tag II for immunochemical detection and the hGH polyadenylation signal sequence. Not containing a transmembrane domain, the nanoluciferase fusion protein is being translocated into the ER and then secreted into the medium. Using a luciferase assay, one can quantifiy the amount of luminescence - and thus, proportionally, the amount of secreted luciferase - by measuring the conversion of luciferin into visible light and integrating it over a timespan of 5 s. Therefore, medium samples were taken every 12 h after transfection of cells and measured via the Promega NanoGlo® luciferase assay system according to the manufacturer's instructions. <br> | ||
| + | |||
| + | [[File:Rsz receptor signalpeptideresults 003.png|thumb|center|850px| '''Figure 3:''' '''A)''' Schematic depiction of the genetic constructs used for signal peptide testing via a secretion luciferase assay. '''B)''' Results of the luciferase assay, showing luminescence in relative luminescence units (RLU) as a function of time for the previously described three different signal peptide constructs as well as a control construct containing no signal peptide. '''C)''' Detection of secreted luciferases in the medium via a Western Blot, using an anti-<i>Strep</i>-tag II antibody as well as an alkaline phosphatase-coupled secondary antibody for detection.]] | ||
| + | |||
| + | ===Discussion: The choice of signal peptide was nailed down=== | ||
| + | As would be expected, the EGFR signal peptide construct showed the lowest level of secretion. Although one might think this could also be attributed to a lower functionality of the signal peptide sequence itself after being translated, the lower secretion level is more likely to stem from lower translation initiation levels. Almost completely missing a 5’ UTR as well as a Kozak sequence, slower translation initiation (being a rate-limiting step) is likely to have lowered the level of translated protein in total. The constructs containing the Igκ and the BM40 signal peptide, on the other hand, contain a longer 5’ UTR as well as the Kozak consensus sequence, and thus result in higher secretion levels. Since the BM40 signal peptide hereby showed the highest secretion levels, it was thus incorporated into the receptor constructs. | ||
| + | <hr> | ||
| − | |||
Latest revision as of 23:45, 19 October 2016
CD4 signal peptide; localizes to plasma membrane
Additional characterization of BBaI712009 by [http://2016.igem.org/Team:LMU-TUM_Munich Munich 2016]:
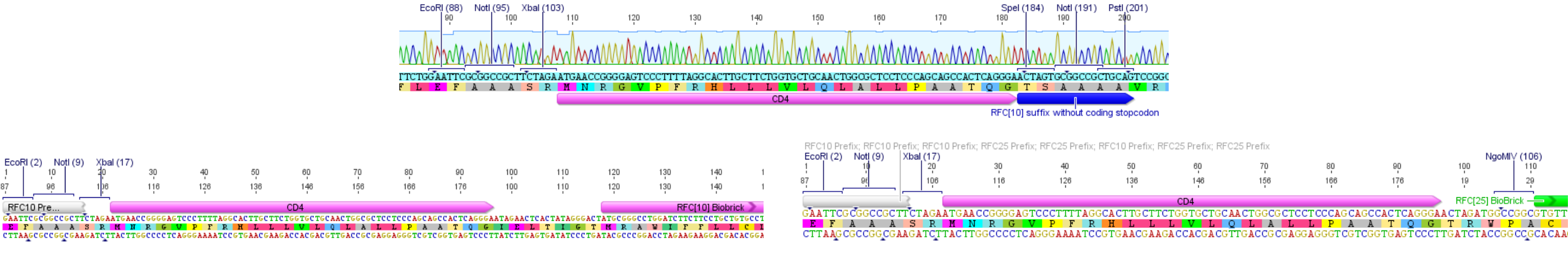
This part was submitted before the RFC[25] standard was established. Thus this part carries a modified RFC[10] pre- and suffix that changes the reading frame of the suffix in a way that, if it is ligated to a second part, it does not generate a stop codon. Although this approach makes it possible to assemble fusion proteins, the ligation to a second part, which does not carry the modified RFC[10], leads to a frameshift in the second part in a way that it is not functional anymore.
Because signal peptides are such important BioBricks we designed new signal peptide parts such as BBa_K2170215 and BBa_K2170214 to help other iGEM teams assembling fusion proteins. The secretion efficiency of the BioBricks was tested by Nano-Glo assay.
Results of signal peptide functional analysis
Bioinformatic signal peptide analysis via the SignalP server
The [http://www.cbs.dtu.dk/services/SignalP SignalP] server, being able to discriminate between the hydrophobic signal peptide sequence and the hydrophobic transmembrane domain[1], can determine whether a sequence possesses the biophysical requirements for functioning as a signal peptide. Furthermore, it is able to predict the probable cleavage site of the signal peptide after its translocation into the ER. For all constructs, signal peptide functionality is predicted, as well as a potential cleavage site. The complete translated amino acid sequences of the respective receptor constructs were used as input. The algorithm for eukaryotes with default D-cutoff value was chosen.

For all three signal peptides, the high S-score indicates high signal peptide functionality. For the Igκ signal peptide, a cleavage site between amino acid 20 and 21 within the signal peptide is predicted. For the BM40 signal peptide, a cleavage site between amino acid 17 and 18 is predicted, and for the EGFR signal peptide, a cleavage site between amino acid 24 and 25 is predicted. </div>
Quantification of signal-peptide mediated translocation via a secretion luciferase assay
For the quantification of signal peptide functionality via a luciferase assay, three constructs were created and tested - each containing one of three signal peptides (the EGFR signal peptide, the Igκ signal peptide or the BM40 signal peptide) and a nanoluciferase as well as a CMV promoter, a Strep-tag II for immunochemical detection and the hGH polyadenylation signal sequence. Not containing a transmembrane domain, the nanoluciferase fusion protein is being translocated into the ER and then secreted into the medium. Using a luciferase assay, one can quantifiy the amount of luminescence - and thus, proportionally, the amount of secreted luciferase - by measuring the conversion of luciferin into visible light and integrating it over a timespan of 5 s. Therefore, medium samples were taken every 12 h after transfection of cells and measured via the Promega NanoGlo® luciferase assay system according to the manufacturer's instructions.
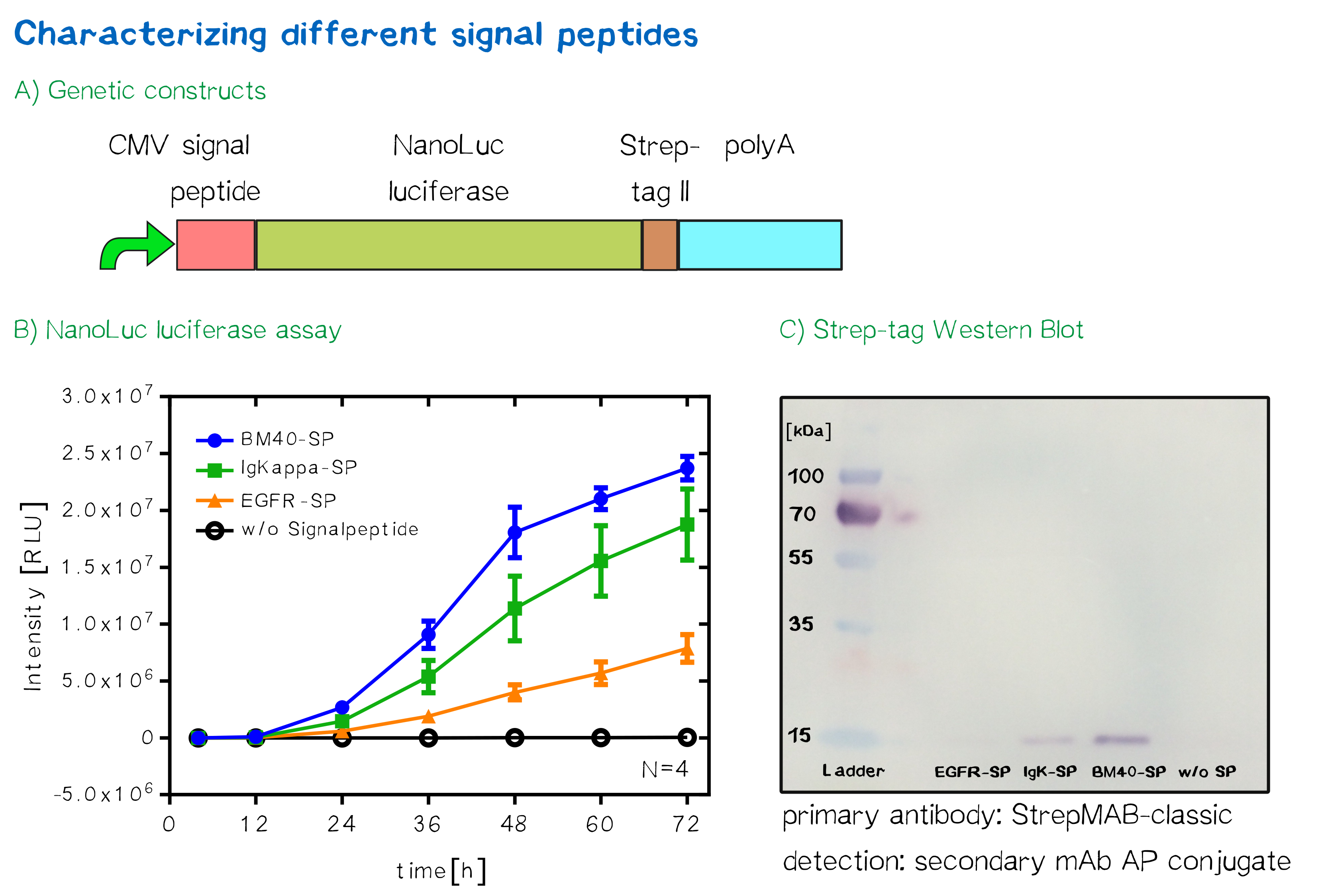
Discussion: The choice of signal peptide was nailed down
As would be expected, the EGFR signal peptide construct showed the lowest level of secretion. Although one might think this could also be attributed to a lower functionality of the signal peptide sequence itself after being translated, the lower secretion level is more likely to stem from lower translation initiation levels. Almost completely missing a 5’ UTR as well as a Kozak sequence, slower translation initiation (being a rate-limiting step) is likely to have lowered the level of translated protein in total. The constructs containing the Igκ and the BM40 signal peptide, on the other hand, contain a longer 5’ UTR as well as the Kozak consensus sequence, and thus result in higher secretion levels. Since the BM40 signal peptide hereby showed the highest secretion levels, it was thus incorporated into the receptor constructs.
Coding region for first 25 amino acids (including start codon) of CD4 receptor which is signal peptide for plasma membrane localization.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
- ↑ Petersen, T. N., Brunak, S., von Heijne, G., & Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature methods, 8(10), 785-786.
