Difference between revisions of "Part:BBa K1899004"
Stephanieyiu (Talk | contribs) |
m |
||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K1899004 short</partinfo> | <partinfo>BBa_K1899004 short</partinfo> | ||
| − | <i>phlFp</i> | + | <i>phlFp</i> - PhlF repressible promoter |
| − | + | <br> | |
| − | + | ||
| + | ==Construct for characterization== | ||
| + | [[File:PhlFp-E0240_2.png|thumb|500px|center|<b>Fig.1 </b> <i>phlFp</i> with reporter gene.]] | ||
| + | In order to compare the promoter strength of the <i>phlFp</i> with the other two promoters, <i>tetp</i> and <i>lacp</i>, it was ligated with a GFP reporter gene, BBa_E0240, in BioBrick RFC10 standard. | ||
===Results=== | ===Results=== | ||
Experiments on comparing the strength of <i>phlFp</i>, <i>tetp</i> and <i>lacp</i>, were performed. Results indicated that <i>phlFp</i> is the strongest among the three, with 1.96 times and 6 times stronger than that of <i>tetp</i> and <i>lacp</i>, respectively. | Experiments on comparing the strength of <i>phlFp</i>, <i>tetp</i> and <i>lacp</i>, were performed. Results indicated that <i>phlFp</i> is the strongest among the three, with 1.96 times and 6 times stronger than that of <i>tetp</i> and <i>lacp</i>, respectively. | ||
| − | + | [[File:IGEM2016 HKUST constructA.png|thumb|600px|center|<b>Fig 2. Comparison on the strength of <i>phlFp</i>, <i>tetp</i> and <i>lacp</i>.</b> Negative control represents BBa_E0240. Characterization was done using <i>E. coli</i> strain JW0336. Cells were first precultured overnight and were subcultured to mid-log phase where GFP emission measurements were made using an EnVision® multilabel reader. This result was obtained by combining 3 characterization data obtained in 3 different days. Error bar present SD from 3 biological replicates.]] | |
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 18: | Line 21: | ||
<partinfo>BBa_K1899004 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1899004 SequenceAndFeatures</partinfo> | ||
| + | ===TetR Orthologs Collection=== | ||
| + | {{Template:Collection/TetR_orthologs/Parts}} | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 15:04, 5 July 2022
phlFp promoter
phlFp - PhlF repressible promoter
Construct for characterization
In order to compare the promoter strength of the phlFp with the other two promoters, tetp and lacp, it was ligated with a GFP reporter gene, BBa_E0240, in BioBrick RFC10 standard.
Results
Experiments on comparing the strength of phlFp, tetp and lacp, were performed. Results indicated that phlFp is the strongest among the three, with 1.96 times and 6 times stronger than that of tetp and lacp, respectively.
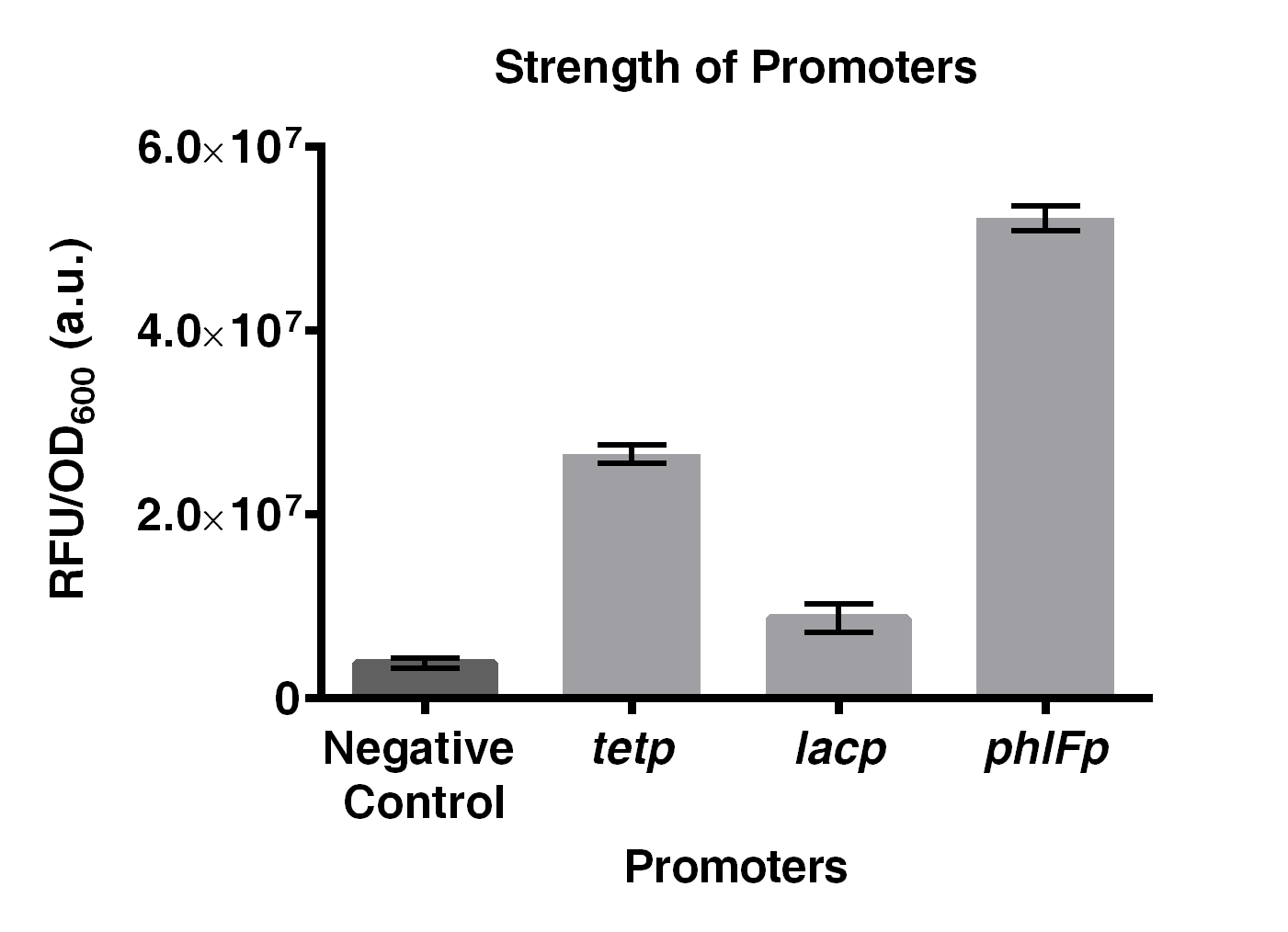
Fig 2. Comparison on the strength of phlFp, tetp and lacp. Negative control represents BBa_E0240. Characterization was done using E. coli strain JW0336. Cells were first precultured overnight and were subcultured to mid-log phase where GFP emission measurements were made using an EnVision® multilabel reader. This result was obtained by combining 3 characterization data obtained in 3 different days. Error bar present SD from 3 biological replicates.
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
TetR Orthologs Collection
| Promoter | CDS | ||
|---|---|---|---|
| pAmeR | BBa_J428000 | AmeR | BBa_J428008 |
| pAmtR | BBa_J428001 | AmtR | BBa_J428009 |
| pBetI | BBa_J428053 | BetI | BBa_J428010 |
| pBM3R1 | BBa_J428054 | BM3R1 | BBa_J428045 |
| pButR | BBa_J428002 | ButR | BBa_J428011 |
| pHapR | BBa_J428003 | HapR | BBa_J428012 |
| pHlyllR | BBa_J428004 | HlyllR | not added |
| pIcaRA | BBa_J428042 | IcaRA | BBa_J428044 |
| pLitR | not added | LitR | BBa_J428043 |
| pLmrA | not added | LmrA | BBa_J428013 |
| Promoter | CDS | ||
|---|---|---|---|
| pMcbR | BBa_J428052 | McbR | BBa_J428014 |
| pOrf2 | not added | Orf2 | BBa_J428015 |
| pPhlF | BBa_K1899004 | PhlF | BBa_J428016 |
| pPsrA | BBa_J428005 | PsrA | BBa_J428017 |
| pQacR | BBa_J428050 | QacR | BBa_J428018 |
| pScbR | BBa_J428006 | ScbR | BBa_J428019 |
| pSmcR | BBa_J428007 | SmcR | not added |
| pSrpR | BBa_J428049 | SrpR | BBa_J428020 |
| pTarA | BBa_J428048 | TarA | BBa_J428021 |

