Difference between revisions of "Part:BBa K1871002"
| (One intermediate revision by the same user not shown) | |||
| Line 21: | Line 21: | ||
This part is the terminator region from yeast alcohol dehydrogenase (ADH1) gene. This stops the RNA polymerase from transcribing the RNA sequence. | This part is the terminator region from yeast alcohol dehydrogenase (ADH1) gene. This stops the RNA polymerase from transcribing the RNA sequence. | ||
| − | =Updated characterization | + | |
| + | |||
| + | =Parts Contribution: Updated characterization added in 2016= | ||
| + | Group: BroadRun-Baltimore 2016 iGEM | ||
| + | Author: BroadRun-Baltimore 2016 Team Members | ||
| + | This part, which was not characterized last year, was re-cloned into a yeast vector, successfully transformed to yeast, and tested, the results of which are below. We were able to prove that this part is functional, and tested in several different conditions. We hope that this improved characterization and confirmation of functionality will help other teams in the future in using this part. | ||
==Proof of Expression and Function== | ==Proof of Expression and Function== | ||
| − | For testing this part, we cloned this composite part into a S.cerevisiae vector | + | For testing this part, we cloned this composite part into a S.cerevisiae vector and transformed this vector with the insert into S.cerevisiae. We combined liquid cultures of our genetically modified S.cerevisae with a starch solution, to determine if the cells were producing the amylase enzyme, and if the enzyme was functioning properly and degrading starch. We found that the cells were indeed producing a functional amylase, evidenced by the significant amount of starch degraded in a period of 6 hours. |
[[File:Brhs_visualstarchdegradation.png|500px]] | [[File:Brhs_visualstarchdegradation.png|500px]] | ||
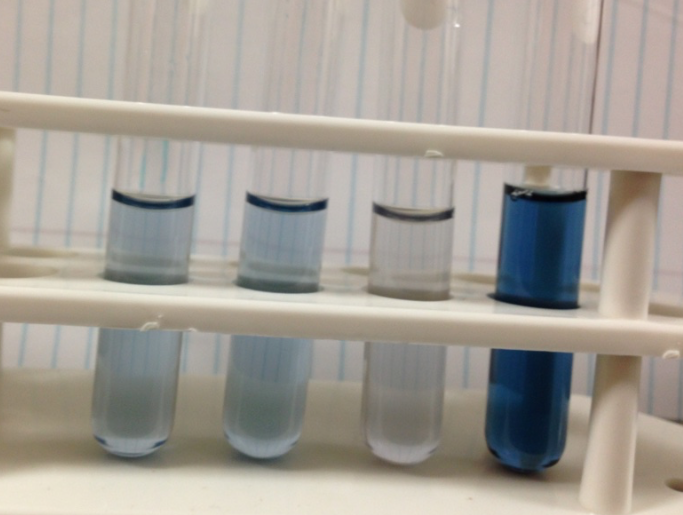
| − | ''Figure 1.'' The image above was taken 5 hours after adding the genetically modified yeast to a starch solution. The yeast strain containing a plasmid with this composite part is the | + | ''Figure 1.'' The image above was taken 5 hours after adding the genetically modified yeast to a starch solution. The yeast strain containing a plasmid with this composite part is the third tube from the left. The right most tube is the control, containing wildtype yeast and starch. Iodine, which reacts with starch to form a blue color, was added to each solution to determine the amount of starch. The third tube from the left has a very light blue color, indicating a small amount of starch left. The right most tube has a dark blue color, indicating a large amount of starch left. This shows that the yeast cells are producing and secreting amylase enzymes, thus the composite part is fully functional. |
[[File:Brhs_graphstarchdegrade.png|500px]] | [[File:Brhs_graphstarchdegrade.png|500px]] | ||
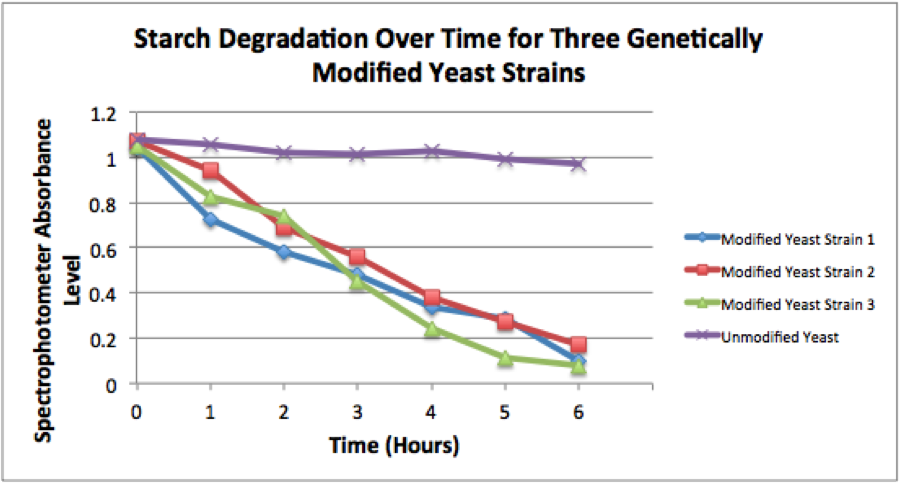
| − | ''Figure 2.'' The image above shows a quantification of starch degradation over a period of 6 hours. A spectrophotometer was used to quantify the amount of blue color in each tube, seen in the figure above. An absorbance level of 1 equals a starch concentration of 0.25%. The yeast strain with this composite part is labeled as "Modified Yeast Strain | + | ''Figure 2.'' The image above shows a quantification of starch degradation over a period of 6 hours. A spectrophotometer was used to quantify the amount of blue color in each tube, seen in the figure above. An absorbance level of 1 equals a starch concentration of 0.25%. The yeast strain with this composite part is labeled as "Modified Yeast Strain 3" in the graph. |
Latest revision as of 15:13, 10 October 2016
Alpha amylase with mating factor alpha secretion
This is the alpha amylase from Bacillus amyloliquefaciens, flanked by Kozak sequence, mating factor alpha secretion sequence, and the ADH1 terminator. This part is controlled by the minimal cyc promoter.
Minimal cyc promoter
This part is the center of the CYC1 promoter, S. cerevisiae. This promoter allows basal transcription, and the transcription signal can be modulated by adding operator sites upstream of this BioBrick. This allows activators or repressors to act on the transcription machinery.
Kozak Sequence
This part initiates translation from eukaryotic mRNA and is cloned between a promoter and coding region to facilitate translation.
Mating Factor Alpha Secretion Sequence
This part is the secretion signal from yeast α-mating factor, and directs the secretion of the produced protein. This allows the exportation of the protein.
Alpha Amylase
Obtained from Bacillus amyloliquefaciens, alpha amylase is a protein enzyme that hydrolyses alpha bonds of large, alpha-linked polysaccharides, such as starch and glycogen.
ADH1 Terminator
This part is the terminator region from yeast alcohol dehydrogenase (ADH1) gene. This stops the RNA polymerase from transcribing the RNA sequence.
Parts Contribution: Updated characterization added in 2016
Group: BroadRun-Baltimore 2016 iGEM Author: BroadRun-Baltimore 2016 Team Members This part, which was not characterized last year, was re-cloned into a yeast vector, successfully transformed to yeast, and tested, the results of which are below. We were able to prove that this part is functional, and tested in several different conditions. We hope that this improved characterization and confirmation of functionality will help other teams in the future in using this part.
Proof of Expression and Function
For testing this part, we cloned this composite part into a S.cerevisiae vector and transformed this vector with the insert into S.cerevisiae. We combined liquid cultures of our genetically modified S.cerevisae with a starch solution, to determine if the cells were producing the amylase enzyme, and if the enzyme was functioning properly and degrading starch. We found that the cells were indeed producing a functional amylase, evidenced by the significant amount of starch degraded in a period of 6 hours.
Figure 1. The image above was taken 5 hours after adding the genetically modified yeast to a starch solution. The yeast strain containing a plasmid with this composite part is the third tube from the left. The right most tube is the control, containing wildtype yeast and starch. Iodine, which reacts with starch to form a blue color, was added to each solution to determine the amount of starch. The third tube from the left has a very light blue color, indicating a small amount of starch left. The right most tube has a dark blue color, indicating a large amount of starch left. This shows that the yeast cells are producing and secreting amylase enzymes, thus the composite part is fully functional.
Figure 2. The image above shows a quantification of starch degradation over a period of 6 hours. A spectrophotometer was used to quantify the amount of blue color in each tube, seen in the figure above. An absorbance level of 1 equals a starch concentration of 0.25%. The yeast strain with this composite part is labeled as "Modified Yeast Strain 3" in the graph.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 105
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1497
Illegal NgoMIV site found at 1512
Illegal AgeI site found at 1815 - 1000COMPATIBLE WITH RFC[1000]