Difference between revisions of "Part:BBa K1806005:Experience"
(→Cloning) |
|||
| (8 intermediate revisions by one other user not shown) | |||
| Line 93: | Line 93: | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| + | <!-- --> | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | ||
| + | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K1806005 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1806005 SequenceAndFeatures</partinfo> | ||
| Line 106: | Line 108: | ||
===User Reviews=== | ===User Reviews=== | ||
| + | ==edit by sysu-medicine=== | ||
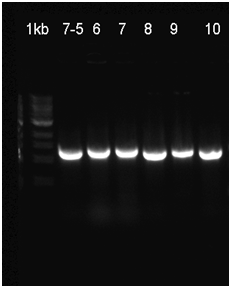
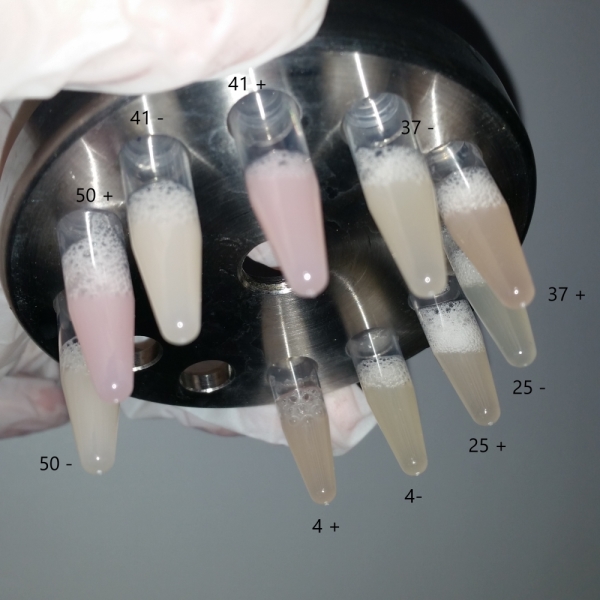
| + | We followed the protocol that had described in this part to repeat some experiment which will show its characterization. We use the restrict enzyme EcoRI and HindIII to cut the plasmids that was extracted from the bacteria solution (Fig.1), and we didn’t have the right DNA stripes which showed in their electrophoresis results. To confirm the plasmid’ s sequence, we also sent the plasmid for sequencing, but we have not received an available result file for its quality score(QS) and continuous reading length (CRL) are under 10. Our bacteria solution cultured in different temperature with or without ITPG had not shown a positive result, and we can’t see any difference of them(Fig.2). | ||
| + | |||
| + | |||
| + | [[file:lyt051.jpg|270px]] | ||
| + | |||
| + | Fig.1 The enzyme cutting product was identified by 1% agarose gel electrophoresis. | ||
| + | |||
| + | |||
| + | [[file:lyt052.jpg|720px]] | ||
| + | |||
| + | Fig.2 The bacteria solution cultured in different temperature. | ||
| + | . | ||
<!-- DON'T DELETE --><partinfo>BBa_K1806005 StartReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_K1806005 StartReviews</partinfo> | ||
<!-- Template for a user review | <!-- Template for a user review | ||
Latest revision as of 01:25, 22 October 2019
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K1806005
Cloning
The already linear part was ligated to the pSB1C3 vector, and then transformated to be verified with Colony PCR. The RFP in the iGEM Registry contained Hind3, unlike the RFP in this part. For this reason, the part was cut with the EcoR1 and Hind3 restriction enzymes. The gel runs of the part were examined. The part was tagged as G-Block 7 and sample 7-6 yielded the accurate base pair length.
Functional Assay
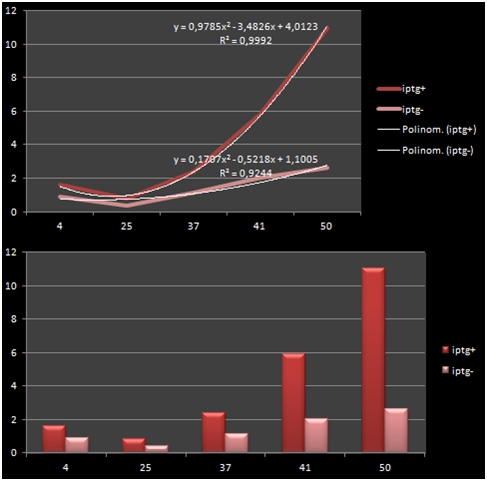
The bacteria that were transformated with G-Block 7 were cultured in 5 ml and incubated for 13 hours. After this incubation, the cultures were incubated at respective temperatures for 4 hours and were given 50uM of iPTG twice with 4 hours periouds. There were a total 5 different temperatures that the cultures were incubated in: 4, 25, 37, 42 and 50 C. The proteins in the cultures were then isolated and the protein concentrations were measured. Data on RFP concentration were acquired at 584 nm of emission and 607 nm of excitation. The acquired values were divided to the total amount of protein to acquire the following ratio.
| Medium Temperature | iPTG Presence +/- | Total Amount of Protein | RFP Fluorometric Measurement | RFP/Total |
|---|---|---|---|---|
| 4 | + | 14.09 | 22.32 | 1.584 |
| 4 | - | 11.349 | 10.36 | 0.912 |
| 25 | + | 17.87 | 14.35 | 0.802 |
| 25 | - | 11.054 | 4,313 | 0,390 |
| 37 | + | 16.808 | 40.2 | 2.391 |
| 37 | - | 15.216 | 17.36 | 1.140 |
| 41 | + | 7.637 | 44.83 | 5.870 |
| 41 | - | 5.557 | 11.13 | 2.002 |
| 50 | + | 5.463 | 60.06 | 10.993 |
| 50 | - | 7.997 | 20.95 | 2.619 |
The graph shows the variance in density concentrations of RFP, caused as a result of the functioning of the iPTG Inducible Promoters in different temperatures. The functioning of the promoters increased cumulatively with increased temperatures.
Usage and Biology
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 2
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 720
Illegal AgeI site found at 832 - 1000COMPATIBLE WITH RFC[1000]
User Reviews
edit by sysu-medicine=
We followed the protocol that had described in this part to repeat some experiment which will show its characterization. We use the restrict enzyme EcoRI and HindIII to cut the plasmids that was extracted from the bacteria solution (Fig.1), and we didn’t have the right DNA stripes which showed in their electrophoresis results. To confirm the plasmid’ s sequence, we also sent the plasmid for sequencing, but we have not received an available result file for its quality score(QS) and continuous reading length (CRL) are under 10. Our bacteria solution cultured in different temperature with or without ITPG had not shown a positive result, and we can’t see any difference of them(Fig.2).
Fig.1 The enzyme cutting product was identified by 1% agarose gel electrophoresis.
Fig.2 The bacteria solution cultured in different temperature. . UNIQ978c61d31348f7ff-partinfo-00000001-QINU UNIQ978c61d31348f7ff-partinfo-00000002-QINU