Difference between revisions of "Part:BBa K1159001:Experience"
(→User Reviews) |
MichaelCWB (Talk | contribs) (→Applications of BBa_K1159001) |
||
| (10 intermediate revisions by 2 users not shown) | |||
| Line 40: | Line 40: | ||
'''IONIS_Paris''' | '''IONIS_Paris''' | ||
<br><br> | <br><br> | ||
| − | <h3> Comparison of the relative light unit | + | |
| + | <h2> Notebook </h2> | ||
| + | <h4>Transformation</h4> | ||
| + | Transformation of competent BL 21(DE3) cells with <bbpart>BBa_K325909</bbpart>, <bbpart>BBa_K1159001</bbpart> and T7-nanoluc plasmids | ||
| + | <br><br> | ||
| + | |||
| + | <h4>Liquid culture of bacteria transformed with pSB1C3-nanoluc (x5) </h4> | ||
| + | <i>Liquid culture on 96 wells plate </i> | ||
| + | When 0,6 < OD<sub>600</sub> < 0,8, induction with arabinose or IPTG Incubate 5h at 30°C; for <bbpart>BBa_K325909</bbpart>, measure the kinetic during the 5h of induction and take the maximum value <br> | ||
| + | Add 50µL of NanoGlow substrate (Promega) for 50 uL of bacteria <br> | ||
| + | Measure kinetic over 120 min and take maximum value for culture with <bbpart>BBa_K1159001</bbpart> and T7-nanoluc | ||
| + | |||
| + | |||
| + | <br><br> | ||
| + | <h2> Results </h2> | ||
| + | <br><br> | ||
| + | <h3> <font style="color:#b22222">Comparison of the relative light unit </font> </h3> | ||
<br> | <br> | ||
| − | <center> [[File:Nanoluc characterization barchart | + | <center> [[File:Nanoluc characterization barchart 2 copie.png|500px]] </center> |
| + | |||
<i><b>Comparison of the relative light unit using 3 differents Biobricks:Induction of BBa_K325909 and BBa_K1159001 with a solution of arabinose 1%, and a solution of IPTG 1 mM for T7-Nanoluc</b></i> | <i><b>Comparison of the relative light unit using 3 differents Biobricks:Induction of BBa_K325909 and BBa_K1159001 with a solution of arabinose 1%, and a solution of IPTG 1 mM for T7-Nanoluc</b></i> | ||
| Line 52: | Line 69: | ||
<br><br> | <br><br> | ||
| − | <h3> Relative Light Unit depending on percentage of arabinose | + | <h3><font style="color:#b22222"> Relative Light Unit depending on percentage of arabinose </font> </h3> |
<br> | <br> | ||
| − | <center> [[File:Nanoluc characterization barchart | + | <center> [[File:Nanoluc characterization barchart 3 copie.png|500px]] </center> |
<br><br> | <br><br> | ||
As mentioned above, we also further characterized the NanoLuc construct of iGEM Munich 2013 by analyzing its expression under different percentage of arabinose. BBa_K1159001 light intensity is linked to the percentage of arabinose. The light intensity increases from 0.01% to 1% of arabinose. However, over 1% of arabinose, the intensity does not increase anymore. | As mentioned above, we also further characterized the NanoLuc construct of iGEM Munich 2013 by analyzing its expression under different percentage of arabinose. BBa_K1159001 light intensity is linked to the percentage of arabinose. The light intensity increases from 0.01% to 1% of arabinose. However, over 1% of arabinose, the intensity does not increase anymore. | ||
| + | |||
| + | ===Applications of BBa_K1159001=== | ||
| + | '''iGEM Marburg 2019''' | ||
| + | <br> | ||
| + | <h2> <b>NanoLuc measured in <i>E. coli</i></b></h2> | ||
| + | In conventional chassis such as E. coli or S.cerevisiae fluorescent proteins are often used as reporter genes to validate genetically modified organisms. However measuring fluorescent activity may not be the best option to choose for phototrophic organisms, because of the absorption caused by the absorption of the photosynthetic pigments of the host organism. | ||
| + | The table below shows the absorption spectra of UTEX 2973. As one can see, reporters such as mTurquoise and YFP underlie a rather high background noise caused by the absorption from the pigments at their ideal wavelength (YFP: 515nm and mTurquoise: 445nm). | ||
| + | |||
| + | https://static.igem.org/mediawiki/parts/3/38/T--Marburg--fluorescent_reporter_spectra_UTEX.png | ||
| + | '''Fig.1-Peak of different fluorescent reporters mapped on the absorption spectra of UTEX2973''' | ||
| + | |||
| + | NanoLuc promises to be a more ideal reporter gene, because there is almost no background noise for this measurement method. To characterize the chosen part from the registry we firstly measured the activity in E.coli before we went any further. | ||
| + | As described in the entry above a dilution series was done to determine the proper dilution for the measurement. After that we mixed 50µl of culture with 50µl of NanoLuc substrate. | ||
| + | Our results were remarkably good, because we needed to dilute the cultures 1:100 in order to get a signal that is under the maximum value, which our plate reader could handle. The results seen below were achieved with cultures grown to OD ~0,8 (600 nm) and the measure points were taken after 39 minutes after treating the cultures with the NanoLuc substrate. | ||
| + | |||
| + | https://2019.igem.org/wiki/images/c/ca/T--Marburg--NanoLuc_measurement_Ecoli.png | ||
| + | '''Fig.2-NanoLuc measurement with 4 biological replicates in E.coli DH5alpha''' | ||
| + | |||
| + | Results: https://2019.igem.org/File:T--Marburg--NanoLuc_measurement_Ecoli_Excel_sheet.docx | ||
===User Reviews=== | ===User Reviews=== | ||
| Line 85: | Line 121: | ||
|width='10%'| | |width='10%'| | ||
<partinfo>BBa_K1159001 AddReview 3</partinfo> | <partinfo>BBa_K1159001 AddReview 3</partinfo> | ||
| − | <I> | + | <I>IONIS_Paris 2015</I> |
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
We observed that no references were performed for the characterization of the BBa_K1159001. That is why, we wanted to compare the first Luxbrick created by Cambridge in 2010, to this first NanoLuc under pBad promoter. BBa_K1159001 showed a light intensity approximately 140 times brighter than BBa_K325909 at equivalent inducer concentration. | We observed that no references were performed for the characterization of the BBa_K1159001. That is why, we wanted to compare the first Luxbrick created by Cambridge in 2010, to this first NanoLuc under pBad promoter. BBa_K1159001 showed a light intensity approximately 140 times brighter than BBa_K325909 at equivalent inducer concentration. | ||
|} | |} | ||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K1159001 AddReview 5</partinfo> | ||
| + | <I>iGEM Marburg 2019</I> | ||
| + | |width='60%' valign='top'| | ||
| + | The part worked nicely and due to its strength is a very good reporter gene. Even after the dilution of 1:100 it showed a 86 fold higher signal than the control. The implementation to our toolbox makes it a perfect fit for phototrophic organisms. | ||
| + | |}; | ||
<!-- DON'T DELETE --><partinfo>BBa_K1159001 EndReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_K1159001 EndReviews</partinfo> | ||
Latest revision as of 23:18, 21 October 2019
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K1159001
iGEM TU Eindhoven 2015
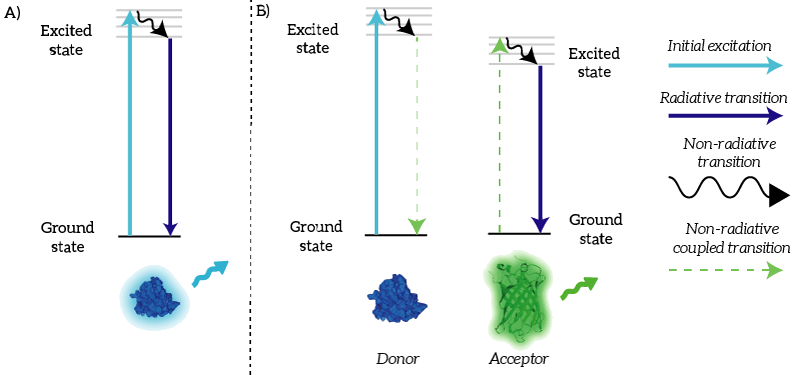
We as iGEM TU Eindhoven 2015 used NanoLuc in a BRET sensor together with mNeonGreen. Bioluminescence Resonance Energy Transfer is a physical process which can take place between fluorophores when they are in close proximity (1-10nm) [1]. An electron which has been transferred to its ‘excited state’ falls back to its ‘ground state’. The energy that is released by the electron falling back to its ground state is normally released in the form of light. In the case of Resonance Energy Transfer, however, the energy of the electron falling back to its ground state is coupled to a transition of an electron in the RET Acceptor from its ground state to the excited state (see Figure 1).
Figure 1: A) Normally, an excited electron falls back to its ground state under the emission of light (a radiative transition). B) In the case of BRET, an excited electron in the donor falls back to its ground state. This transition is coupled to the excitation of an electron in the acceptor. This excited electron falls back normally under the emission of light.
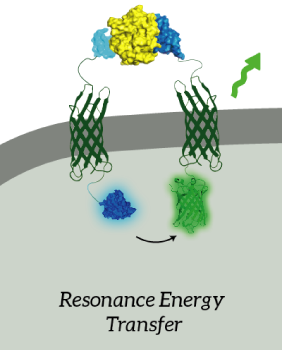
The main goal of our project was to design a universal, modular biosensor that could detect molecules by using aptamers. NanoLuc worked very well for us. We've inserted NanoLuc in a pETDuet-1 vector together with Outer Membrane Protein X (OmpX) and a BsoBI-linker, which is a long and flexible GGSGGS-linker with two BsoBI restriction sites. Several experiments gave very strong results for NanoLuc. Together with an OmpX-mNeonGreen construct, BRET can take place between the two fluorophores.
Figure 2: The OmpX-NanoLuc construct together with the OmpX-mNeonGreen construct. When in close proximity, BRET takes places from NanoLuc to mNeonGreen.
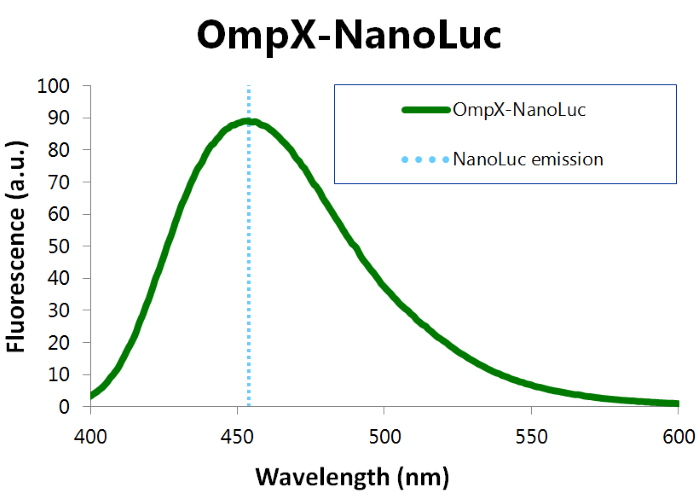
We've characterized NanoLuc by performing a bioluminescence assay with the OmpX-NanoLuc construct. This construct was inserted in the pET-Duet-1 vector, which has a T7-promoter. IPTG induces this T7-promotor. The bioluminescence assay gave the following results (see Figure 3).
Figure 3: Bioluminescence results of our OmpX-NanoLuc construct.
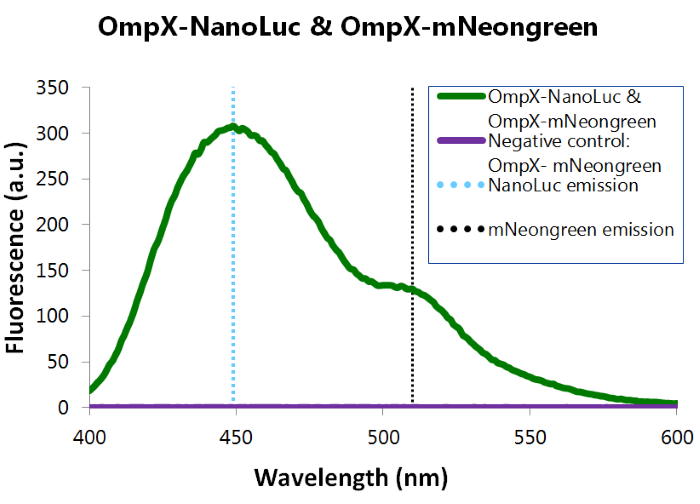
We've also performed a bioluminescence assay for the construct that both contains OmpX-NanoLuc and OmpX-mNeonGreen. This gave the following results (see Figure 4). From this it can be concluded that both the construct were present, that NanoLuc worked well as a BRET donor and that mNeonGreen worked well as a BRET acceptor.
Figure 4: Bioluminescence results of our complete construct, containing both NanoLuc and mNeonGreen.
References:
Medintz I. and Hildebrandt N., Eds., FRET - Förster Resonance Energy Transfer. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA, 2013.
Applications of BBa_K1159001
IONIS_Paris
Notebook
Transformation
Transformation of competent BL 21(DE3) cells with BBa_K325909, BBa_K1159001 and T7-nanoluc plasmids
Liquid culture of bacteria transformed with pSB1C3-nanoluc (x5)
Liquid culture on 96 wells plate
When 0,6 < OD600 < 0,8, induction with arabinose or IPTG Incubate 5h at 30°C; for BBa_K325909, measure the kinetic during the 5h of induction and take the maximum value
Add 50µL of NanoGlow substrate (Promega) for 50 uL of bacteria
Measure kinetic over 120 min and take maximum value for culture with BBa_K1159001 and T7-nanoluc
Results
Comparison of the relative light unit
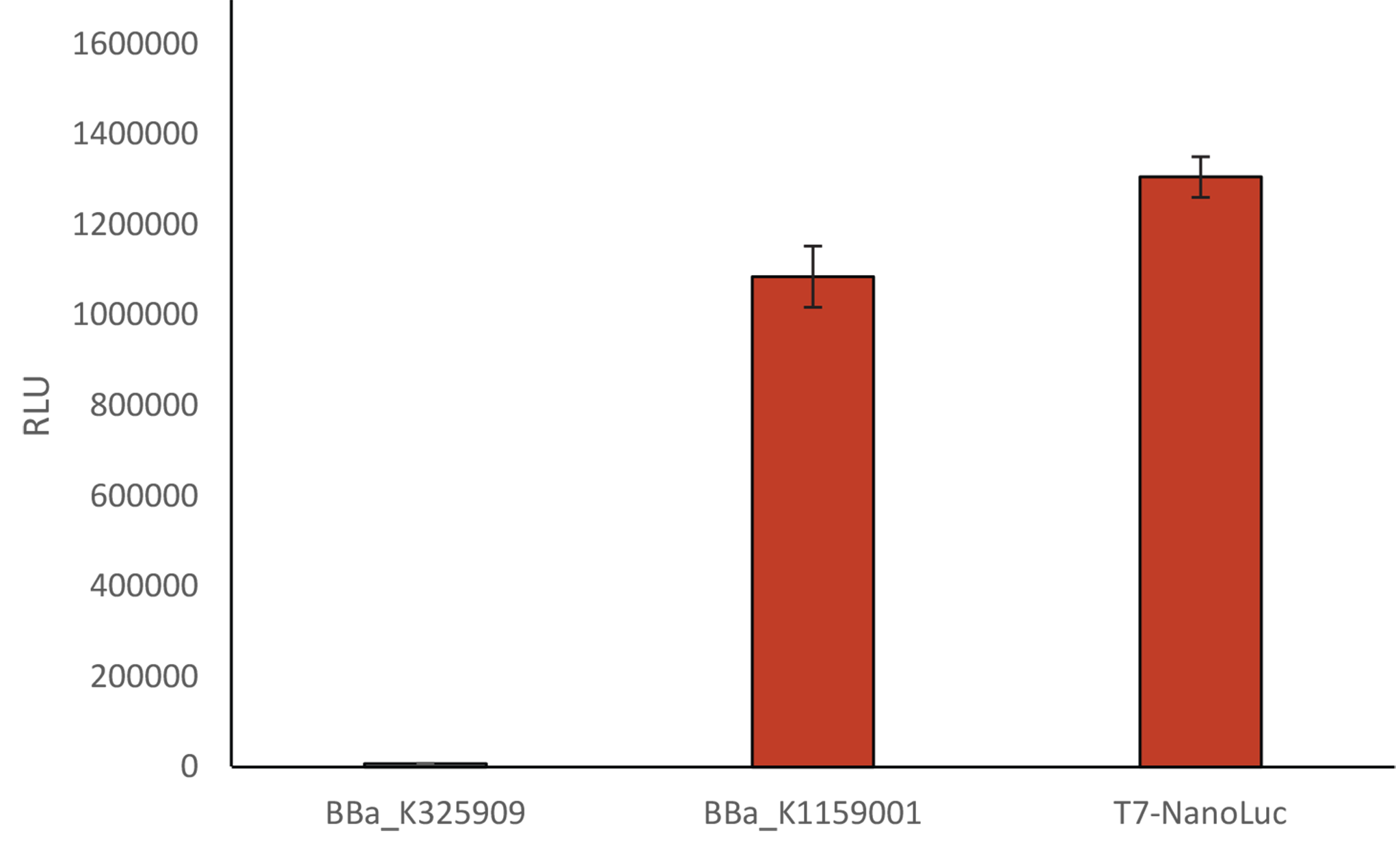
Comparison of the relative light unit using 3 differents Biobricks:Induction of BBa_K325909 and BBa_K1159001 with a solution of arabinose 1%, and a solution of IPTG 1 mM for T7-Nanoluc
We compared our new T7 Nanoluc to the first Luxbrick created by Cambridge in 2010 (BBa_K325909), and to BBa_K1159001. T7 NanoLuc showed a 1,2 times brighter light intensity (RLU) than BBa_K1159001 and a light intensity approximately 140 times brighter than BBa_K325909.
Relative Light Unit depending on percentage of arabinose
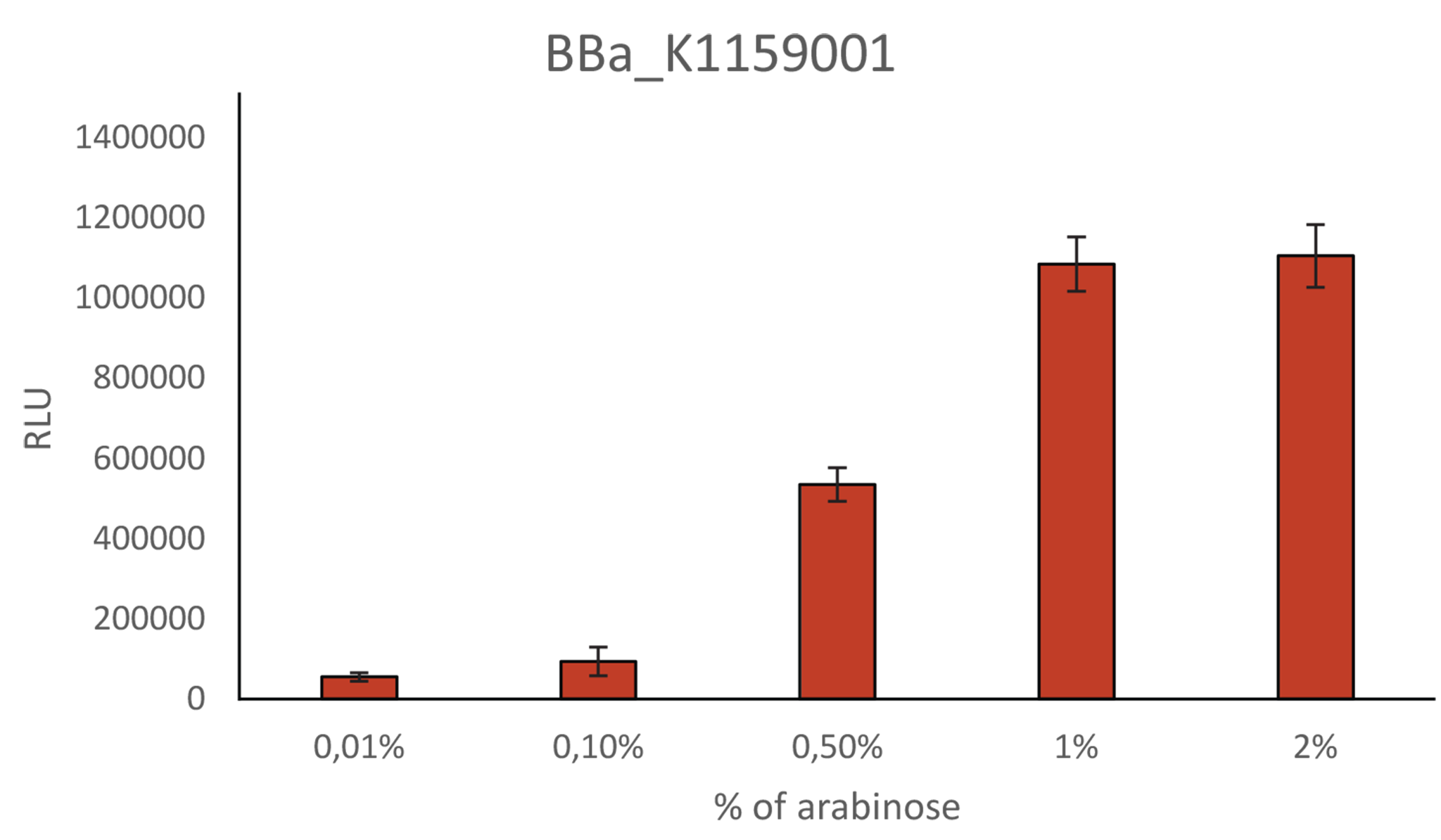
As mentioned above, we also further characterized the NanoLuc construct of iGEM Munich 2013 by analyzing its expression under different percentage of arabinose. BBa_K1159001 light intensity is linked to the percentage of arabinose. The light intensity increases from 0.01% to 1% of arabinose. However, over 1% of arabinose, the intensity does not increase anymore.
Applications of BBa_K1159001
iGEM Marburg 2019
NanoLuc measured in E. coli
In conventional chassis such as E. coli or S.cerevisiae fluorescent proteins are often used as reporter genes to validate genetically modified organisms. However measuring fluorescent activity may not be the best option to choose for phototrophic organisms, because of the absorption caused by the absorption of the photosynthetic pigments of the host organism. The table below shows the absorption spectra of UTEX 2973. As one can see, reporters such as mTurquoise and YFP underlie a rather high background noise caused by the absorption from the pigments at their ideal wavelength (YFP: 515nm and mTurquoise: 445nm).
 Fig.1-Peak of different fluorescent reporters mapped on the absorption spectra of UTEX2973
Fig.1-Peak of different fluorescent reporters mapped on the absorption spectra of UTEX2973
NanoLuc promises to be a more ideal reporter gene, because there is almost no background noise for this measurement method. To characterize the chosen part from the registry we firstly measured the activity in E.coli before we went any further. As described in the entry above a dilution series was done to determine the proper dilution for the measurement. After that we mixed 50µl of culture with 50µl of NanoLuc substrate. Our results were remarkably good, because we needed to dilute the cultures 1:100 in order to get a signal that is under the maximum value, which our plate reader could handle. The results seen below were achieved with cultures grown to OD ~0,8 (600 nm) and the measure points were taken after 39 minutes after treating the cultures with the NanoLuc substrate.
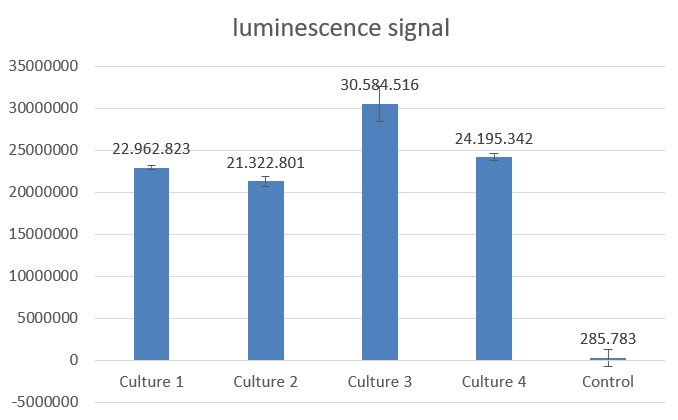 Fig.2-NanoLuc measurement with 4 biological replicates in E.coli DH5alpha
Fig.2-NanoLuc measurement with 4 biological replicates in E.coli DH5alpha
Results: https://2019.igem.org/File:T--Marburg--NanoLuc_measurement_Ecoli_Excel_sheet.docx
User Reviews
UNIQ32753c4bf114861e-partinfo-00000004-QINU
|
•••••
iGEM TU Eindhoven 2015 |
This part works very well. We used NanoLuc in a BRET sensor, connected to OmpX with a BsoBI-linker. When furimazine is added, NanoLuc will work as a donor, which means that the energy of an electron falling back to its ground state is coupled to a transition of an electron in the RET Acceptor from its ground state to the excited state. We used mNeonGreen as acceptor in this BRET sensor. For more information and further characterization, see the Applications section above. |
|
•••
IONIS_Paris 2015 |
We observed that no references were performed for the characterization of the BBa_K1159001. That is why, we wanted to compare the first Luxbrick created by Cambridge in 2010, to this first NanoLuc under pBad promoter. BBa_K1159001 showed a light intensity approximately 140 times brighter than BBa_K325909 at equivalent inducer concentration. |
|
•••••
iGEM Marburg 2019 |
The part worked nicely and due to its strength is a very good reporter gene. Even after the dilution of 1:100 it showed a 86 fold higher signal than the control. The implementation to our toolbox makes it a perfect fit for phototrophic organisms. |
UNIQ32753c4bf114861e-partinfo-00000008-QINU