Difference between revisions of "Part:BBa K1639000"
(→Characterization) |
|||
| (9 intermediate revisions by the same user not shown) | |||
| Line 12: | Line 12: | ||
A common limitation for riboregulators has been their dynamic range (Liu et al., 2012). Previous prokaryotic translational riboregulators have typically modulated biological signals by up to a maximum of 55-fold for activators (Callura et al., 2012) and up to 10-fold for repressors (Mutalik et al., 2012). In contrast, protein-based transcriptional regulators have demonstrated dynamic ranges over an order of magnitude higher, with widely-used inducible promoters regulating protein expression over 350-fold (Lutz and Bujard, 1997) and sigma factor-promoter pairs providing up to 480-fold modulation (Rhodius et al., 2013).Despite the inherent programmability of RNA-based systems, efforts at constructing large sets of orthogonal riboregulators have been limited to libraries of at most seven parts with crosstalk levels of 20% (Takahashi and Lucks, 2013). Typical RNA-based regulators employ interaction domains consisting of30 nts, which corresponds to a sequence space of over 1018 potential regulatory elements. Thus, the sheer diversity of possible RNA-based parts suggests that previous devices have not come close to realizing the potential of highly orthogonal regulation. | A common limitation for riboregulators has been their dynamic range (Liu et al., 2012). Previous prokaryotic translational riboregulators have typically modulated biological signals by up to a maximum of 55-fold for activators (Callura et al., 2012) and up to 10-fold for repressors (Mutalik et al., 2012). In contrast, protein-based transcriptional regulators have demonstrated dynamic ranges over an order of magnitude higher, with widely-used inducible promoters regulating protein expression over 350-fold (Lutz and Bujard, 1997) and sigma factor-promoter pairs providing up to 480-fold modulation (Rhodius et al., 2013).Despite the inherent programmability of RNA-based systems, efforts at constructing large sets of orthogonal riboregulators have been limited to libraries of at most seven parts with crosstalk levels of 20% (Takahashi and Lucks, 2013). Typical RNA-based regulators employ interaction domains consisting of30 nts, which corresponds to a sequence space of over 1018 potential regulatory elements. Thus, the sheer diversity of possible RNA-based parts suggests that previous devices have not come close to realizing the potential of highly orthogonal regulation. | ||
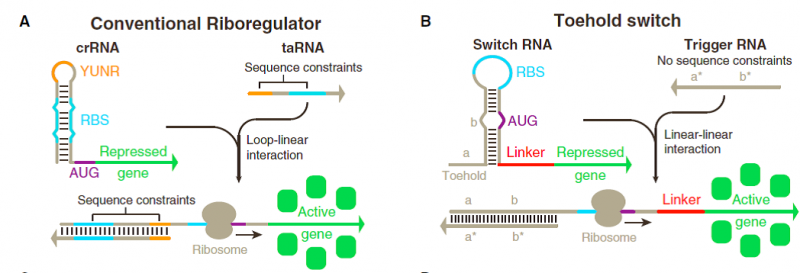
| − | [[File:ATOMS-Turkiye_ulcer_andgate_1.5.png]] | + | [[File:ATOMS-Turkiye_ulcer_andgate_1.5.png|600px|thumb|left|'''Figure 5:''' (A and B) Design schematics of conventional riboregulators (A) and toehold switches (B). Variable sequences are shown in gray, whereas conserved or constrained sequences are represented by different colors.]] |
Much of this discrepancy arises from the significant sequence constraints imposed on riboregulators engineered thus far (Figure1A). Like natural riboregulators, engineered riboregulators of translation have invariably used base pairing to the ribosome binding site (RBS) to prevent ribosome binding, thereby preventing translation (Callura et al., 2012; Isaacs et al., 2004; Mutaliket al., 2012; Rodrigo et al., 2012). Because repression is caused by RBS binding, trigger RNAs that activate translation are engineered to contain an RBS sequence to displace the repressing sequence, which in turn reduces the potential sequence space for the riboregulator. | Much of this discrepancy arises from the significant sequence constraints imposed on riboregulators engineered thus far (Figure1A). Like natural riboregulators, engineered riboregulators of translation have invariably used base pairing to the ribosome binding site (RBS) to prevent ribosome binding, thereby preventing translation (Callura et al., 2012; Isaacs et al., 2004; Mutaliket al., 2012; Rodrigo et al., 2012). Because repression is caused by RBS binding, trigger RNAs that activate translation are engineered to contain an RBS sequence to displace the repressing sequence, which in turn reduces the potential sequence space for the riboregulator. | ||
| Line 22: | Line 22: | ||
Toehold switch systems are composed of two RNA strands referred to as the switch and trigger (Figure 1B). The switch RNA contains the coding sequence of the gene being regulated. Upstream of this coding sequence is a hairpin-based processing module containing both a strong RBS and a start codon that is followed by a common 21 nt linker sequence coding for low-molecular-weight amino acids added to the N terminus of the gene of interest. A single-stranded toehold sequence at the 50 end of the hairpin module provides the initial binding site for the trigger RNA strand. This trigger molecule contains an extended single stranded region that completes a branch migration process with the hairpin to expose the RBS and start codon, thereby initiating translation of the gene of interest. The hairpin processing unit functions as a repressor of translation in the absence of the trigger strand. Unlike previous riboregulators, the RBS sequence is left completely unpaired within the 11 nt loop of the hairpin. Instead, the bases immediately before and after the start codon are sequestered within RNA duplexes that are 6 bp and 9 bp long, respectively. The start codon itself is left unpaired in the switches we tested, leaving a 3 nt bulge near the midpoint of the 18 nt hairpin stem. Because the repressing domain b (Figure 1B) does not possess complementary bases to the start codon, the cognate trigger strand in turn does not need to contain corresponding start codon bases, thereby increasing the number of potential trigger sequences. The sequence in the hairpin added after the start codon was also screened for the presence of stop codons, as they would prematurely terminate translation of the gene of interest when the riboregulator was activated. We employed a 12 nt Toehold domain at the 50 end of the hairpin to initiate its interaction with the cognate trigger strand. The trigger RNA contains a 30 nt single-stranded RNA sequence that is complementary to the toehold and stem of the switch RNA. | Toehold switch systems are composed of two RNA strands referred to as the switch and trigger (Figure 1B). The switch RNA contains the coding sequence of the gene being regulated. Upstream of this coding sequence is a hairpin-based processing module containing both a strong RBS and a start codon that is followed by a common 21 nt linker sequence coding for low-molecular-weight amino acids added to the N terminus of the gene of interest. A single-stranded toehold sequence at the 50 end of the hairpin module provides the initial binding site for the trigger RNA strand. This trigger molecule contains an extended single stranded region that completes a branch migration process with the hairpin to expose the RBS and start codon, thereby initiating translation of the gene of interest. The hairpin processing unit functions as a repressor of translation in the absence of the trigger strand. Unlike previous riboregulators, the RBS sequence is left completely unpaired within the 11 nt loop of the hairpin. Instead, the bases immediately before and after the start codon are sequestered within RNA duplexes that are 6 bp and 9 bp long, respectively. The start codon itself is left unpaired in the switches we tested, leaving a 3 nt bulge near the midpoint of the 18 nt hairpin stem. Because the repressing domain b (Figure 1B) does not possess complementary bases to the start codon, the cognate trigger strand in turn does not need to contain corresponding start codon bases, thereby increasing the number of potential trigger sequences. The sequence in the hairpin added after the start codon was also screened for the presence of stop codons, as they would prematurely terminate translation of the gene of interest when the riboregulator was activated. We employed a 12 nt Toehold domain at the 50 end of the hairpin to initiate its interaction with the cognate trigger strand. The trigger RNA contains a 30 nt single-stranded RNA sequence that is complementary to the toehold and stem of the switch RNA. | ||
| − | === | + | ===Characterization=== |
PSB1C3-Trigger RNA/pColA-Toehold-GFP Cotransformation: | PSB1C3-Trigger RNA/pColA-Toehold-GFP Cotransformation: | ||
| Line 32: | Line 32: | ||
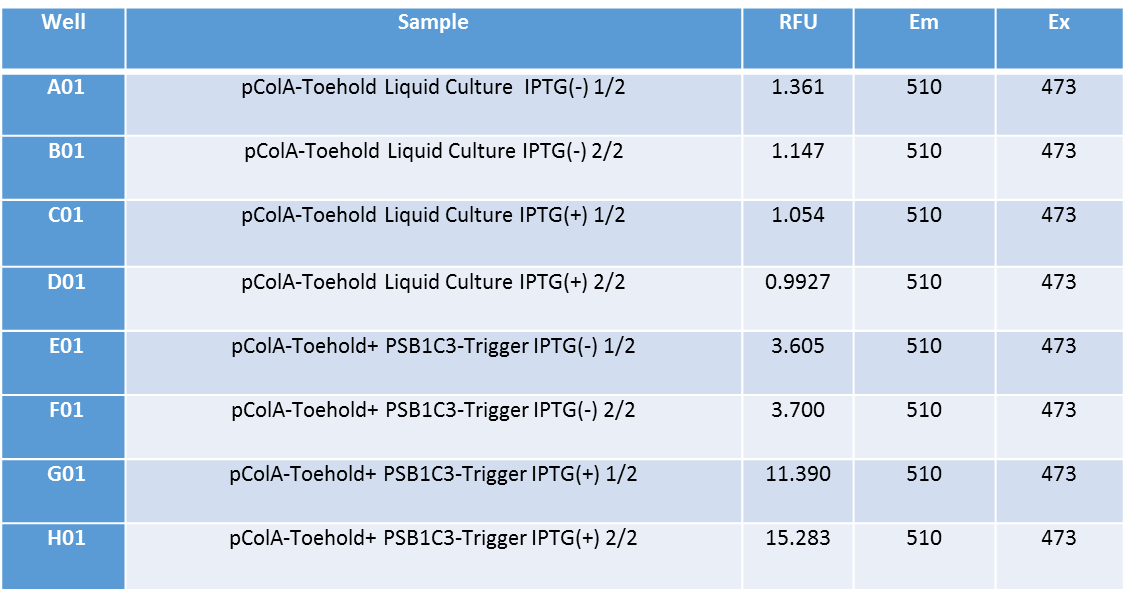
For the purposes given above, we made liquid culture of two different bacteria together; bacteria including only pColA-Toehold-GFP plasmid and bacteria including pCola-Toehold-GFP and PSB1C3-Trigger RNA. We incubated these bacteria in liquid culture for 13 hours at 37C and added IPTG into their mediums at 13. hour. After adding IPTG, we incubated them for more 3 hours at 37C. At the end of three hours, we firstly measured GFP fluorescence of the grown cultures by using VarioScan device. The results of measurements are given above. They were made twice. | For the purposes given above, we made liquid culture of two different bacteria together; bacteria including only pColA-Toehold-GFP plasmid and bacteria including pCola-Toehold-GFP and PSB1C3-Trigger RNA. We incubated these bacteria in liquid culture for 13 hours at 37C and added IPTG into their mediums at 13. hour. After adding IPTG, we incubated them for more 3 hours at 37C. At the end of three hours, we firstly measured GFP fluorescence of the grown cultures by using VarioScan device. The results of measurements are given above. They were made twice. | ||
| − | [[File:ATOMS-Turkiye_ulcer_andgate_3.r8.png| | + | [[File:ATOMS-Turkiye_ulcer_andgate_3.r8.png|480px|thumb|left|'''Figure 2''']] |
| − | + | [[File:ATOMS-Turkiye_ulcer_andgate_3.2.png|400px|thumb|right|'''Figure 3''']] | |
| − | [[File:ATOMS-Turkiye_ulcer_andgate_3.2.png| | + | <br clear=all> |
When the results are analyzed, it is obvious that Toehold-Trigger RNA parts gave very high amounts of flourescence together while Toehold-GFP part gave flourescence in a very low amount. There is almost 15 times difference between these two fluoroscence amounts. This proves that Toehold-Trigger system works successfully. Also it is shown that systems work IPTG-dependently. | When the results are analyzed, it is obvious that Toehold-Trigger RNA parts gave very high amounts of flourescence together while Toehold-GFP part gave flourescence in a very low amount. There is almost 15 times difference between these two fluoroscence amounts. This proves that Toehold-Trigger system works successfully. Also it is shown that systems work IPTG-dependently. | ||
| Line 40: | Line 40: | ||
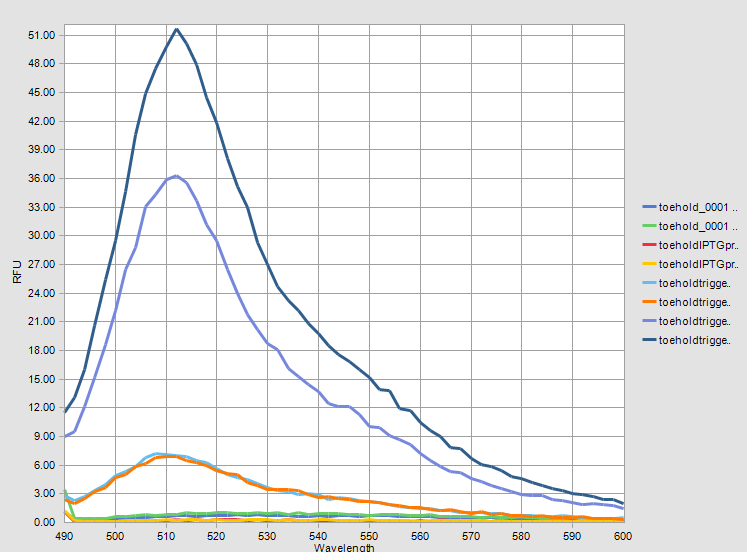
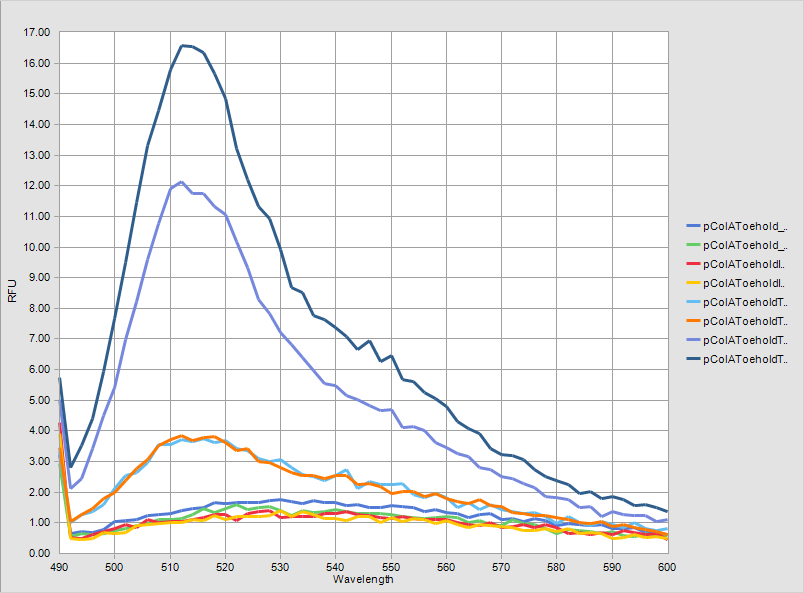
The graphs of measurement results are given below. The difference between two systems can be observed clearly. | The graphs of measurement results are given below. The difference between two systems can be observed clearly. | ||
| − | [[File:ATOMS-Turkiye_ulcer_andgate_3.r9.png| | + | [[File:ATOMS-Turkiye_ulcer_andgate_3.r9.png|600px|thumb|center|'''Figure 4:''' GFP flourescent measurrement in LB mediums.]] |
| + | |||
We also observed the liquid cultures under the fluorescence microscope. | We also observed the liquid cultures under the fluorescence microscope. | ||
| − | [[File:ATOMS-Turkiye_ulcer_andgate_3.3.png|900px|]] | + | [[File:ATOMS-Turkiye_ulcer_andgate_3.3.png|900px|thumb|'''Figure 5:''' Flourescence microscope image of GFP producing E. coli.]] |
| − | + | For better results, we isolated protein and we fluorimetrically measured them. Firstly we centrifuged the liquid cultures which were incubated for 16 hours. We took pictures of tubes after centrifugation, they are shown belown. | |
| − | [ | + | [[File:ATOMS-Turkiye_ulcer_andgate_3.4.png|600px|center|thumb|'''Figure 6:''' Pellets can be seen after 16 hours of liquid culter precipitated.]] |
| − | |||
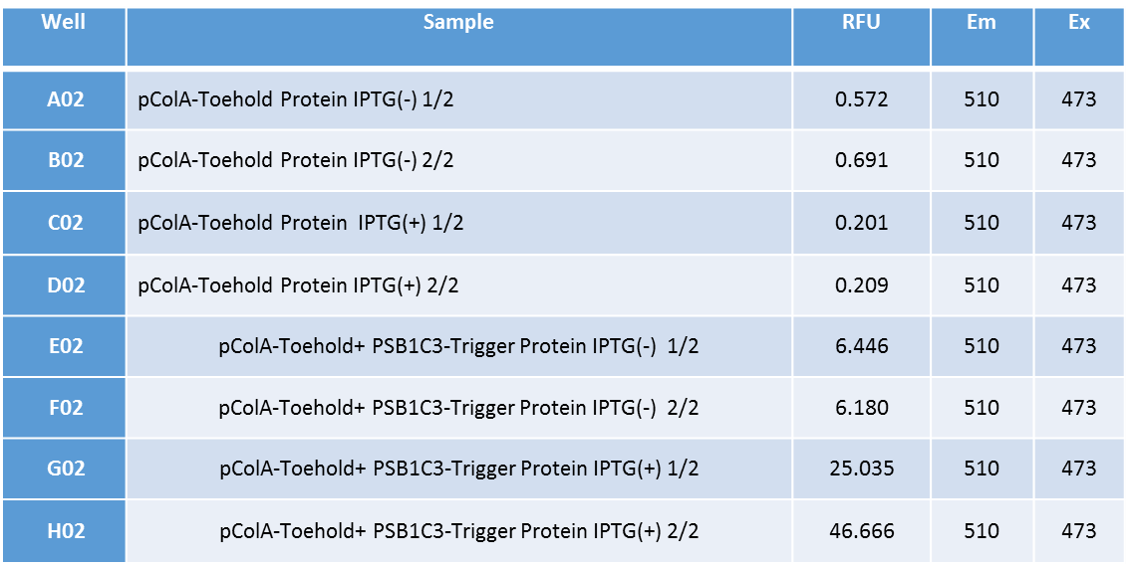
| − | + | We applied Standard protein isolation protocol after this centrifuge and managed to isolate protein from the occured pellets. We made GFP fluorescence measurement with VarioScan from those isolated proteins. The results of measurements are shown below. | |
| − | [ | + | [[File:ATOMS-Turkiye_ulcer_andgate_3.r10.png|480px|thumb|left|]] |
| + | [[File:ATOMS-Turkiye_ulcer_andgate_3.5.png|400px|thumb|right|]] | ||
| + | <br clear=all> | ||
| − | + | The measurement results of isolated proteins indicate that Toehold-Trigger system works very efficiently. GFP fluorescence amount of Toehold-trigger RNA including bacteria is about 175 times more than only Toehold including bacteria’s. also the leak of this system is almost negligible. In the absence of Trigger RNA, Toehold’s GFP fluorescence is almost zero. This indicates that our system works very efficient and in a very specific way. | |
| − | [ | + | [[File:ATOMS-Turkiye_ulcer_andgate_3.6.png|600px|center|thumb|'''Figure 9:''' Protein extract GFP florasans measurement]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
| Line 87: | Line 72: | ||
<partinfo>BBa_K1639000 parameters</partinfo> | <partinfo>BBa_K1639000 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | ===Sources=== | ||
| + | |||
| + | Green, A., Silver, P., Collins, J., & Yin, P. (n.d.). Toehold Switches: De-Novo-Designed Regulators of Gene Expression. Cell, 925-939. | ||
Latest revision as of 21:28, 20 September 2015
Toehold-GFP
Toehold-GFP reporter device. This part produces reporter flourescence protein GFP, only when a specific mRNA present in the cell called Trigger RNA(BBa_K1639001).
Usage and Biology
Toehold switches provide a high level of orthogonality and can be forward engineered to provide average dynamic range above 400. Toehold switches, with their wide dynamic range, orthogonality, and programmability, represent a versatile and powerful platform for regulation of translation, offering diverse applications in molecular biology, synthetic biology, and biotechnology.New classes of regulatory components that offer wide dynamic range, low system crosstalk, and design flexibility represent a much-needed, enabling step toward fully realizing the potential of synthetic biology in areas such as biotechnology and medicine. (Khalil and Collins, 2010).
Engineered riboregulators consist of cognate pairs of RNAs: a transducer strand that regulates translation or transcription and a trans-acting RNA that binds to the transducer to modulate its biological activity. Riboregulator designs can be classified according to the initial RNA-RNA interaction that drives hybridization between the transducer and trans-acting RNAs. Reactions initiated between loop sequences in both RNAs are termed loop-loop interactions, whereas those that occur between a loop sequence and an unstructured RNA are termed loop-linear(Takahashi and Lucks, 2013).
A common limitation for riboregulators has been their dynamic range (Liu et al., 2012). Previous prokaryotic translational riboregulators have typically modulated biological signals by up to a maximum of 55-fold for activators (Callura et al., 2012) and up to 10-fold for repressors (Mutalik et al., 2012). In contrast, protein-based transcriptional regulators have demonstrated dynamic ranges over an order of magnitude higher, with widely-used inducible promoters regulating protein expression over 350-fold (Lutz and Bujard, 1997) and sigma factor-promoter pairs providing up to 480-fold modulation (Rhodius et al., 2013).Despite the inherent programmability of RNA-based systems, efforts at constructing large sets of orthogonal riboregulators have been limited to libraries of at most seven parts with crosstalk levels of 20% (Takahashi and Lucks, 2013). Typical RNA-based regulators employ interaction domains consisting of30 nts, which corresponds to a sequence space of over 1018 potential regulatory elements. Thus, the sheer diversity of possible RNA-based parts suggests that previous devices have not come close to realizing the potential of highly orthogonal regulation.
Much of this discrepancy arises from the significant sequence constraints imposed on riboregulators engineered thus far (Figure1A). Like natural riboregulators, engineered riboregulators of translation have invariably used base pairing to the ribosome binding site (RBS) to prevent ribosome binding, thereby preventing translation (Callura et al., 2012; Isaacs et al., 2004; Mutaliket al., 2012; Rodrigo et al., 2012). Because repression is caused by RBS binding, trigger RNAs that activate translation are engineered to contain an RBS sequence to displace the repressing sequence, which in turn reduces the potential sequence space for the riboregulator.
Previous riboregulators have also relied on U-turn loop structures to drive loop-loop and loop-linear interactions between RNAs (Figure 1A) (Callura et al., 2012; Isaacs et al., 2004; Luckset al., 2011; Takahashi and Lucks, 2013). U-turn loops are common RNA structural motifs formed by tertiary interactions that have been identified in ribozymes, ribosomal RNAs, and transfer RNA anticodon loops (Gutell et al., 2000). Although recent work has begun to show that loops with canonical U-turn sequences are not essential for riboregulators (Mutalik et al., 2012; Rodrigoet al., 2012), the engineered systems reported to date have continued their reliance on the loop-mediated RNA interactions from natural systems. Although these loop interactions have been selected by evolution in nature, alternative approaches employing linear-linear RNA interactions are amenable to rational engineering and exhibit more favorable reaction kinetics and thermodynamics, factors that could be exploited to increase riboregulator dynamic range.
A riboregulator that activates gene expression must switch from a secondary structure that prevents translation to a configuration that promotes translation upon binding of a cognate trans-acting RNA. Although the Shine-Dalgarno sequence is an important factor in determining the efficiency of translation from a given mRNA, studies have found that secondary structure in regions near by the start codon also plays a critical role (Kudla et al., 2009).Furthermore, genome-wide analyses have revealed strong biases toward low secondary structures around the start codon of mRNAs from a panel of hundreds of bacterial genomes.
Toehold switch systems are composed of two RNA strands referred to as the switch and trigger (Figure 1B). The switch RNA contains the coding sequence of the gene being regulated. Upstream of this coding sequence is a hairpin-based processing module containing both a strong RBS and a start codon that is followed by a common 21 nt linker sequence coding for low-molecular-weight amino acids added to the N terminus of the gene of interest. A single-stranded toehold sequence at the 50 end of the hairpin module provides the initial binding site for the trigger RNA strand. This trigger molecule contains an extended single stranded region that completes a branch migration process with the hairpin to expose the RBS and start codon, thereby initiating translation of the gene of interest. The hairpin processing unit functions as a repressor of translation in the absence of the trigger strand. Unlike previous riboregulators, the RBS sequence is left completely unpaired within the 11 nt loop of the hairpin. Instead, the bases immediately before and after the start codon are sequestered within RNA duplexes that are 6 bp and 9 bp long, respectively. The start codon itself is left unpaired in the switches we tested, leaving a 3 nt bulge near the midpoint of the 18 nt hairpin stem. Because the repressing domain b (Figure 1B) does not possess complementary bases to the start codon, the cognate trigger strand in turn does not need to contain corresponding start codon bases, thereby increasing the number of potential trigger sequences. The sequence in the hairpin added after the start codon was also screened for the presence of stop codons, as they would prematurely terminate translation of the gene of interest when the riboregulator was activated. We employed a 12 nt Toehold domain at the 50 end of the hairpin to initiate its interaction with the cognate trigger strand. The trigger RNA contains a 30 nt single-stranded RNA sequence that is complementary to the toehold and stem of the switch RNA.
Characterization
PSB1C3-Trigger RNA/pColA-Toehold-GFP Cotransformation: To show And Gate system works properly, these two plasmids with different origins should be transformed into the same bacteria. T7 is the promoter which produces Trigger RNA and Toehold-GFP mRNA’s , so we assured that they were cotransformed into a bacteria strain including T7 RNA polymerase. BL21 served this purpose, and after cotransformation, we observed grown colonies.
Functional Assay: We designed a functional assay setup in order to figure out if Toehold-Trigger RNA system is functional. The main goal of this setup is to show that; Toehold-GFP doesn’t give fluorecence alone but if it comes together with Trigger RNA, GFP fluorescences. GFP production is also IPTG-dependent.
For the purposes given above, we made liquid culture of two different bacteria together; bacteria including only pColA-Toehold-GFP plasmid and bacteria including pCola-Toehold-GFP and PSB1C3-Trigger RNA. We incubated these bacteria in liquid culture for 13 hours at 37C and added IPTG into their mediums at 13. hour. After adding IPTG, we incubated them for more 3 hours at 37C. At the end of three hours, we firstly measured GFP fluorescence of the grown cultures by using VarioScan device. The results of measurements are given above. They were made twice.
When the results are analyzed, it is obvious that Toehold-Trigger RNA parts gave very high amounts of flourescence together while Toehold-GFP part gave flourescence in a very low amount. There is almost 15 times difference between these two fluoroscence amounts. This proves that Toehold-Trigger system works successfully. Also it is shown that systems work IPTG-dependently.
The graphs of measurement results are given below. The difference between two systems can be observed clearly.
We also observed the liquid cultures under the fluorescence microscope.
For better results, we isolated protein and we fluorimetrically measured them. Firstly we centrifuged the liquid cultures which were incubated for 16 hours. We took pictures of tubes after centrifugation, they are shown belown.
We applied Standard protein isolation protocol after this centrifuge and managed to isolate protein from the occured pellets. We made GFP fluorescence measurement with VarioScan from those isolated proteins. The results of measurements are shown below.
The measurement results of isolated proteins indicate that Toehold-Trigger system works very efficiently. GFP fluorescence amount of Toehold-trigger RNA including bacteria is about 175 times more than only Toehold including bacteria’s. also the leak of this system is almost negligible. In the absence of Trigger RNA, Toehold’s GFP fluorescence is almost zero. This indicates that our system works very efficient and in a very specific way.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 117
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 772
Sources
Green, A., Silver, P., Collins, J., & Yin, P. (n.d.). Toehold Switches: De-Novo-Designed Regulators of Gene Expression. Cell, 925-939.