Difference between revisions of "Part:BBa K1694006"
| (7 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1694006 short</partinfo> | <partinfo>BBa_K1694006 short</partinfo> | ||
<h1>'''Introduction'''</h1> | <h1>'''Introduction'''</h1> | ||
| − | [[File:GBPpart.png|200px|thumb|right|'''Fig.1'''Gold Binding Polypeptide]] | + | [[File:GBPpart.png|200px|thumb|right|'''Fig.1''' Gold Binding Polypeptide]] |
| − | The gold binding polypeptide, abbreviated as GBP, is | + | The gold binding polypeptide, abbreviated as GBP, is composed of three amino acid sequence repetition of MHGKTQATSGTIQS. This ''E. coli'' cell-surface display system has developed for several years<sup>[2]</sup>. The recent computation study demonstrated that multiple repeats of GBP have higher affinity to gold surface, because a high degree of conformational flexibility can make the binding much stronger. |
| − | The | + | The connection mechanism between GBP and gold metal plane is still unknown. By using the concept of '''molecular dynamics''' (MD), GBP, with the structure of antiparallel β-sheet, is shown to attach to gold surface via OH-binding. It seems that the hydroxyl, as well as the amine ligands on GBP, can recognize the atomic lattice structure of gold for the alignment of the molecule along the variants of a six-fold axis on the Au (111) surface<sup>[3]</sup>. In addition, another study also states that water molecules played an important role in the surface recognition by the side chain and hydration layer of the peptides to reach the better binding <sup>[4]</sup>.<br> |
''Reference:<br>'' | ''Reference:<br>'' | ||
| Line 14: | Line 14: | ||
<h1>'''Experiment'''</h1> | <h1>'''Experiment'''</h1> | ||
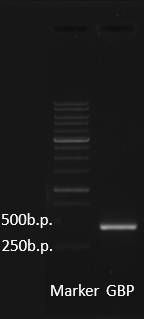
[[File:GBP.png|left|thumb|900px|<p style="padding: 10px !important;">'''Fig.2''' The PCR result of the GBP. The DNA sequence length of GBP are around 100~200 bp, so the PCR products should appear at 300~400 bp.</p>]] | [[File:GBP.png|left|thumb|900px|<p style="padding: 10px !important;">'''Fig.2''' The PCR result of the GBP. The DNA sequence length of GBP are around 100~200 bp, so the PCR products should appear at 300~400 bp.</p>]] | ||
| − | After receiving the DNA sequences from the gene synthesis company, we recombined each GBP gene to | + | After receiving the DNA sequences from the gene synthesis company, we recombined each GBP gene to pSB1C3 backbones and conducted a PCR experiment to check the size of each of the GBP. The DNA sequence length of the GBP is around 100~150 bp. In this PCR experiment, the GBP products size should be near at 350~400 bp. The Fig.2 showed the correct size of the GBP, and proved that we successful ligated the GBP sequence onto an ideal backbone. |
[[File:EXGBP.png|600px|thumb|center|'''Fig.3'''Plate of gold binding polypeptide]] | [[File:EXGBP.png|600px|thumb|center|'''Fig.3'''Plate of gold binding polypeptide]] | ||
<br> | <br> | ||
<br> | <br> | ||
| + | |||
| + | <h1>'''Application'''</h1> | ||
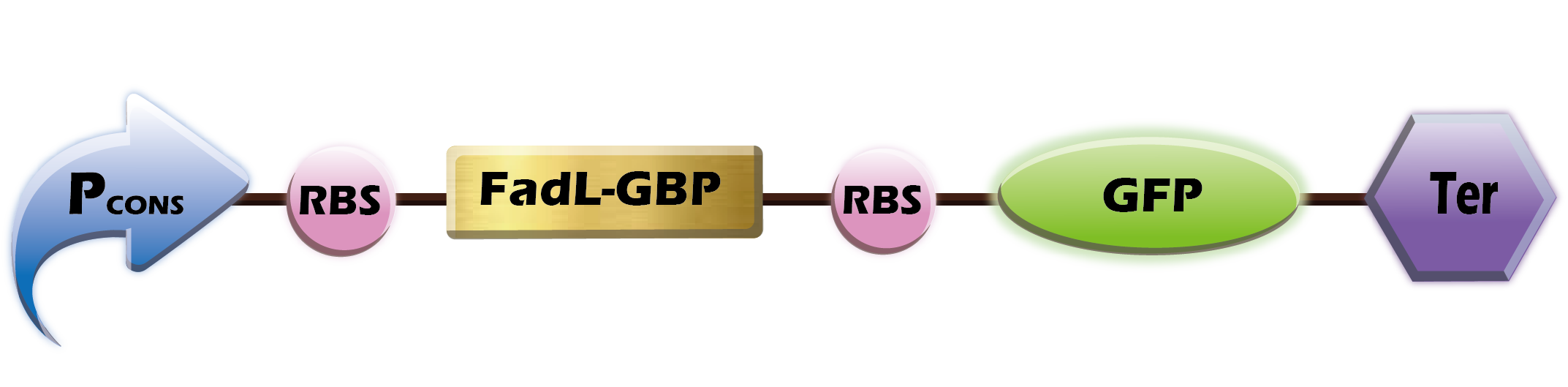
| + | Our team wants to display GBP on the surface of ''E. coli'', so we create the FadL-GBP part.<br> | ||
| + | |||
| + | <p style="font-size:120%">'''FadL'''</p> | ||
| + | The FadL protein (48.8 kDa) is an outer membrane protein of the ''E. coli'', which plays an important role in the uptake of exogenous long-chain fatty acid. The FadL protein has twenty antiparallel β-strands, which form a β-barrel structure and are connected by 9 internal loops and 10 external loops. Due to the β-structure, the FadL protein is able to span the outer membrane multiple times to form a long-chain fatty acid-specific channel. With these properties, the truncated FadL protein can be used as a novel anchoring motif for display of proteins on the ''E. coli'' cell surface<sup> [1]</sup>. | ||
| + | |||
| + | ''Reference:''<br> | ||
| + | ''[1]Display of Bacterial Lipase on the Escherichia coli Cell Surface by Using FadL as an Anchoring Motif and Use of the Enzyme in Enantioselective Biocatalysis, Seung Hwan Lee1, Jong-Il Choi1, Si Jae Park1,†, Sang Yup Lee1,2,* and Byoung Chul Park3(2004)'' | ||
| + | |||
| + | [[File:GBPGFP.png|600px|thumb|center|'''Fig.4'''Pcons+B0034+FadL-GBP+B0030+GFP+Ter]] | ||
| + | |||
| + | |||
| + | We intended to make the observation of the result more directly by producing green fluorescence at the same time. To make the comparison of the binding efficiency, we selected the genetic sequence, Pcons + RBS +GFP +Ter, as the negative control group. Because there was no existence of fully-functioned phenomenon, there was no positive control in our experiment. | ||
| + | |||
| + | <div style="display: block; height:300pt;"> | ||
| + | [[File:GBP-a.png|400px|thumb|left|'''Fig.5(a)'''the results of the test group, which contains the genetic sequence of Pcons+ B0034 + FadL-GBP + B0030 + GFP + Ter]] | ||
| + | [[File:GBPb.png|400px|thumb|left|'''Fig.5(b)'''displays the consequence of the control group with the genetic sequence, Pcon+ RBS + GFP + Ter ]] | ||
| + | </div> | ||
| + | |||
| + | According to the pictures shown above, we can see that there is more ''E. coli'' with gold-binding polypeptide being observed on the gold chip. It indicates that the ''E. coli'' with gold-binding polypeptide can attach on gold chip more effectively and efficiently than others without gold-binding polypeptide. | ||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 07:57, 22 September 2015
Gold Binding Polypeptide
Introduction
The gold binding polypeptide, abbreviated as GBP, is composed of three amino acid sequence repetition of MHGKTQATSGTIQS. This E. coli cell-surface display system has developed for several years[2]. The recent computation study demonstrated that multiple repeats of GBP have higher affinity to gold surface, because a high degree of conformational flexibility can make the binding much stronger.
The connection mechanism between GBP and gold metal plane is still unknown. By using the concept of molecular dynamics (MD), GBP, with the structure of antiparallel β-sheet, is shown to attach to gold surface via OH-binding. It seems that the hydroxyl, as well as the amine ligands on GBP, can recognize the atomic lattice structure of gold for the alignment of the molecule along the variants of a six-fold axis on the Au (111) surface[3]. In addition, another study also states that water molecules played an important role in the surface recognition by the side chain and hydration layer of the peptides to reach the better binding [4].
Reference:
[1] Molecular characterization of a prokaryotic polypeptide sequence that catalyzes Au crystal formation, John L. Kulp III,a Mehmet Sarikayab and John Spencer Evans, Journal of Materials Chemistry(2004)
[2] Assembly of Gold-Binding Proteins for Biomolecular Recognition, Zareie HM1,2* and Sarikaya M3, Austin Journal of Biosensors & Bioelectronics (2015)
[3] Thermodynamics of Engineered Gold Binding Peptides: Establishing the Structure−Activity Relationships, Urartu Ozgur Safak Seker, Brandon Wilson, John L. Kulp,§ John S. Evans, Candan Tamerler, and Mehmet Sarikaya(2014)
Experiment
After receiving the DNA sequences from the gene synthesis company, we recombined each GBP gene to pSB1C3 backbones and conducted a PCR experiment to check the size of each of the GBP. The DNA sequence length of the GBP is around 100~150 bp. In this PCR experiment, the GBP products size should be near at 350~400 bp. The Fig.2 showed the correct size of the GBP, and proved that we successful ligated the GBP sequence onto an ideal backbone.
Application
Our team wants to display GBP on the surface of E. coli, so we create the FadL-GBP part.
FadL
The FadL protein (48.8 kDa) is an outer membrane protein of the E. coli, which plays an important role in the uptake of exogenous long-chain fatty acid. The FadL protein has twenty antiparallel β-strands, which form a β-barrel structure and are connected by 9 internal loops and 10 external loops. Due to the β-structure, the FadL protein is able to span the outer membrane multiple times to form a long-chain fatty acid-specific channel. With these properties, the truncated FadL protein can be used as a novel anchoring motif for display of proteins on the E. coli cell surface [1].
Reference:
[1]Display of Bacterial Lipase on the Escherichia coli Cell Surface by Using FadL as an Anchoring Motif and Use of the Enzyme in Enantioselective Biocatalysis, Seung Hwan Lee1, Jong-Il Choi1, Si Jae Park1,†, Sang Yup Lee1,2,* and Byoung Chul Park3(2004)
We intended to make the observation of the result more directly by producing green fluorescence at the same time. To make the comparison of the binding efficiency, we selected the genetic sequence, Pcons + RBS +GFP +Ter, as the negative control group. Because there was no existence of fully-functioned phenomenon, there was no positive control in our experiment.
According to the pictures shown above, we can see that there is more E. coli with gold-binding polypeptide being observed on the gold chip. It indicates that the E. coli with gold-binding polypeptide can attach on gold chip more effectively and efficiently than others without gold-binding polypeptide.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]