Difference between revisions of "Part:BBa K629003:Experience"
(→Applications of BBa_K629003) |
(→Applications of BBa_K629003) |
||
| Line 24: | Line 24: | ||
| − | + | </i> | |
<B>UT-Tokyo 2015 characterized BBa_K629003 !</B> | <B>UT-Tokyo 2015 characterized BBa_K629003 !</B> | ||
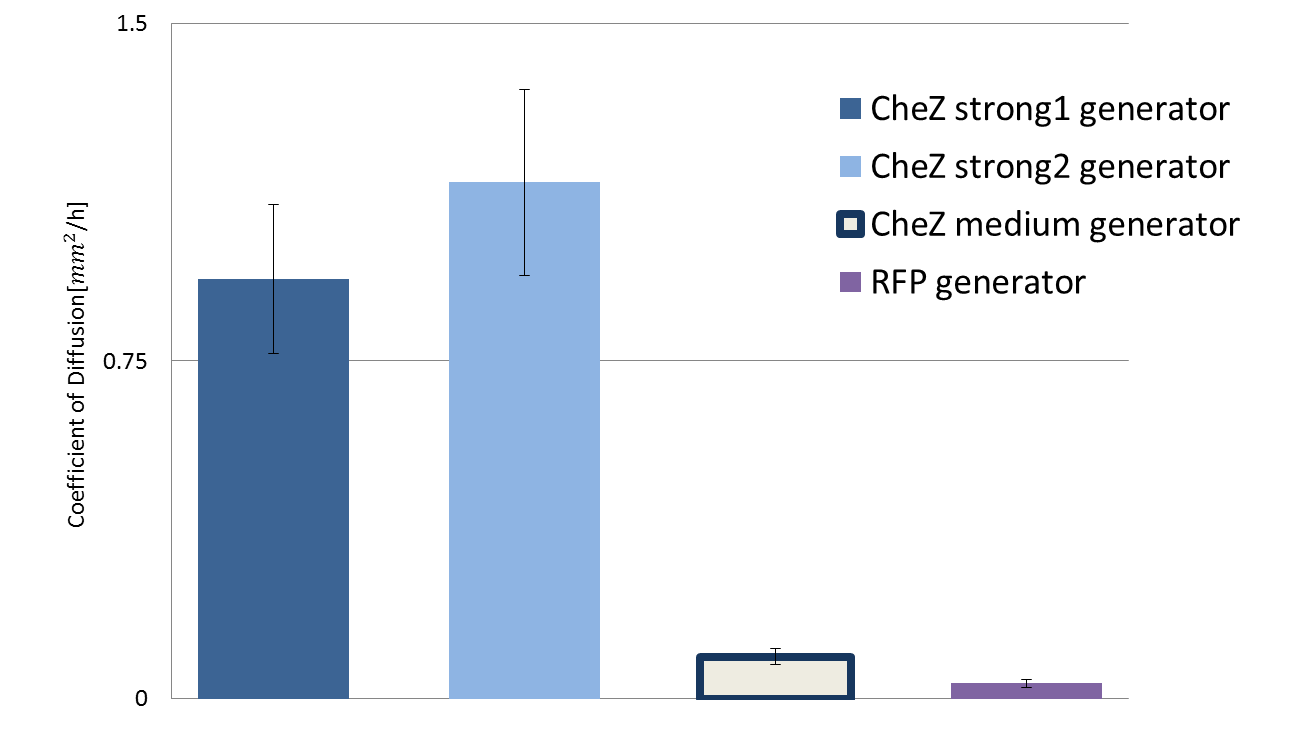
<p>We constructed new composite CheZ generators([[Part:BBa_K1631018]], [[part:BBa_K1631019]], [[part:BBa_K1631020]]) and introduced them to CheZ knockout strain named JW1870 (Keio collection). In contrast to previous iGEM projects, we tried to measure diffusion rate of <i>E.coli</i> quantitavely. In this end we took time lapse photography to measure the colony expanding speed. By combination with measurement of growth rate we could successfully quantitate diffusion rates according to different strength of CheZ | <p>We constructed new composite CheZ generators([[Part:BBa_K1631018]], [[part:BBa_K1631019]], [[part:BBa_K1631020]]) and introduced them to CheZ knockout strain named JW1870 (Keio collection). In contrast to previous iGEM projects, we tried to measure diffusion rate of <i>E.coli</i> quantitavely. In this end we took time lapse photography to measure the colony expanding speed. By combination with measurement of growth rate we could successfully quantitate diffusion rates according to different strength of CheZ | ||
Latest revision as of 23:58, 18 September 2015
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K629003
Restoring chemotaxis
Uppsala iGEM 2014 did characterization experiments to see if chemotaxis could be restored in a mutant strain, UU2685, of E.coli with cheZ knocked out. The results were compared to a motile strain of E.coli, RP437. Both strains were obtained from Prof. Parkinson. To test the motility swarming plate assay was conducted. To improve the preformance and consistency of the swarmplates, incubation was done in room temperature instead of 37 degrees celcius. Read more about the assay on our [http://2014.igem.org/Team:Uppsala/Project_Targeting wiki].

Figure 1. Swarmplate assays incubated for 2 days in room temperature
1)positive control (RP437 with empty pSB1C3 plasmid) [220]
2)negative control (UU2685 with empty pSB1C3 plasmid) [216]
3)B0034_J23100-cheZ (UU2685) [170]
4)B0034_J23113-cheZ (UU2685) [171]
5)B0034_J23114-cheZ (UU2685) [172]
On all plates presented in figure 2, positive controls (strain 1) swarmed, while negative controls (strain 2) did not swarm. The altered protocol for the swarm plate assay was thus able to distinguish motile strains from non-motile strains.
Strain 3, containing a cheZ expression cassette with the J23100 promoter, swarmed all six times when it was tested, though not to the same extent as the positive control. Strain 4, containing a cheZ expression cassette with the J23113 promoter, also showed to be motile, although it only swarmed to a small extent.
The result of the strain containing the cheZ expression cassette including the J23114 promoter (strain 5) is not as clear as that of the other two constructs. The structures that have grown upon incubation resemble the negative control. For strain 5, these structures include cloudy formations, however, which is most clearly visible in figure 2C.
UT-Tokyo 2015 characterized BBa_K629003 !
We constructed new composite CheZ generators(Part:BBa_K1631018, part:BBa_K1631019, part:BBa_K1631020) and introduced them to CheZ knockout strain named JW1870 (Keio collection). In contrast to previous iGEM projects, we tried to measure diffusion rate of E.coli quantitavely. In this end we took time lapse photography to measure the colony expanding speed. By combination with measurement of growth rate we could successfully quantitate diffusion rates according to different strength of CheZ expression.
User Reviews
UNIQ7de07c93bb539366-partinfo-00000000-QINU UNIQ7de07c93bb539366-partinfo-00000001-QINU