Difference between revisions of "Part:BBa K401001"
(→Improvements) |
Cccxyyyyyyyy (Talk | contribs) (→Sequence) |
||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 14: | Line 14: | ||
The second part consists of the GRE followed by the reporter gene. In our experiments, we used LacZ for this purpose. The amplification of this part could not be made because of some problems found when trying to find the sequence of the GRE. | The second part consists of the GRE followed by the reporter gene. In our experiments, we used LacZ for this purpose. The amplification of this part could not be made because of some problems found when trying to find the sequence of the GRE. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Sup35p== | ==Sup35p== | ||
| Line 102: | Line 59: | ||
==Improvements== | ==Improvements== | ||
The 2015 [http://2015.igem.org/Team:Kent Kent] iGEM team improved this BioBrick by removing all illegal cutting sites within the coding region to improve compatibility with the BioBrick standard. . Kent produced Sup35NM BioBricks, one containing only Sup35 (Part name: [https://parts.igem.org/wiki/index.php?title=Part:BBa_K1739000 BBa_K1739000]) and one containing both Sup35NM and the CsgAss signal sequence that targets the protein for export (Part name: [https://parts.igem.org/wiki/index.php?title=Part:BBa_K1739002 BBa_K1739002]). | The 2015 [http://2015.igem.org/Team:Kent Kent] iGEM team improved this BioBrick by removing all illegal cutting sites within the coding region to improve compatibility with the BioBrick standard. . Kent produced Sup35NM BioBricks, one containing only Sup35 (Part name: [https://parts.igem.org/wiki/index.php?title=Part:BBa_K1739000 BBa_K1739000]) and one containing both Sup35NM and the CsgAss signal sequence that targets the protein for export (Part name: [https://parts.igem.org/wiki/index.php?title=Part:BBa_K1739002 BBa_K1739002]). | ||
| + | |||
==References== | ==References== | ||
Latest revision as of 17:47, 26 October 2017
NMGR526 (construction)
Introduction
We got to send to the registry the first biobrick composed by the coding sequence of a protein with a potential prion behaviour. The capability of using a constitutive transcription activator whose function is dependent of this particular mechanism can be very useful in many different conditions. The system allows the simultaneous presence of two phenotypes, conferring an intrinsic diversity in the population that can lead to an evolutionary advantage or to a very interesting dynamics.
Components of the prionic switch
The switch is formed by two different parts: the activator and the reporter. The activator part is a construction of two fragments: the NM domains of the protein Sup35, which confers to the protein the prionic activity, and the GR526 portion, which contains the DNA-binding and transcription-activation domains. The ligand-binding domain of the GR (Glucocorticoid Receptor) protein was eliminated, decoupling the response of the protein to the presence of glucocorticoids, and thus generating a constitutive transcription activation factor. The normal activity of this protein results in the activation of the genes preceded by the GRE (Glucocorticoid Response Element). When exposed to heat shock or other stressful conditions, the NM domains activation to a prionic conformation is induced, eventually inhibiting the activation of transcription.
This part was amplified by using the primers indicated by Li and Lindquist (2000), together with the sequence recommended to use for the ligation protocol with the plasmid pSB1C3. Those primers are:
- Forward actagtagcggccgctgcagATGTCGGATTCAAAC
- Reverse tctagaagcggccgcgaattcTCCTGCAGTGGCTTG
(again, capital letters represent the region that pairs with the coding sequence of NMGR526).
The second part consists of the GRE followed by the reporter gene. In our experiments, we used LacZ for this purpose. The amplification of this part could not be made because of some problems found when trying to find the sequence of the GRE.
Sup35p
[PSI+] is a non-Mendelian trait of Saccharomyces cerevisae that supress nonsense codons. This phenotype is due to a self-replication conformation (prion state) of a protein encoded by the gene Sup35. This protein, Sup35p, is the yeast eukariotyc release factor 3 (eRF3) and forms the translation termination complex with Sup45p (eRF1). The function of Sup35p is releasing the nascent polypeptide chain from the ribosome through GTP hydrolysis when Sup45p recognize a stop codon (Shorter and Lindquist, 2005).
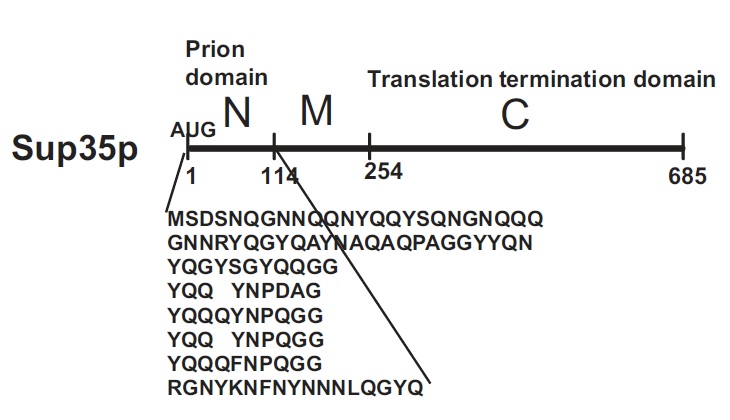
Sup35p is 685 amino acids long and has three distinct parts (Fig.2). The NH2-terminus (N) is termed the prion-forming domain (PrD) because plays a critical role in Sup35p’s changes in proteic conformation and it is responsible for its prion behaviour. This domain is 114 amino acids long and has a high content in glutamine and asparagine. The middle region (M) provides a solubilizing and/or spacing function. Finally, the COOH-terminus (C) is responsible for the translation-termination activity.
In [PSI+] cells, most Sup35p is insoluble and nonfunctional, causing an increase in the translational read-through of stop codons. This trait is heritable because Sup35p in the amyloid state as every prion influences new Sup35p to adopt the same conformation and passes from mother cell to daughter. In [psi-] cells, the translation-termination factor Sup35p is soluble and functional.
Behaviour
Sup35p is a subunit of the translation termination complex. Its prionic nature has been proposed to have some effect on the stress response, as a possible mechanism to obtain modified genetic expression products. When the prionic conformation is activated, the termination of translation is less effective and thus new longer proteins form (True and Lindquist, 2000; Tyedmers et al., 2008). When the sequence corresponding to the NM domains of Sup35p is fused to other gene, the protein resulting of this construction acquires the prionic behaviour (Li and Lindquist, 2000).
On the other side, GR activates the transcription of genes preceded by GRE (Glucocorticoid Receptor Element) when steroid hormones are present (Heitzer et al., 2007). However, it becomes a constitutive transcription activator when it lacks its C terminal ligand-binding domain (Schena and Yamamoto, 1988). Because of the length of amino acids of the cut protein, this short version of GR is named GR526.
Li and Lindquist (2000) showed that the fused protein (NMGR526) is a functional constitutive transcription activator. In addition, when the prionic conformation is reached because of the presence of a certain stimulus, NMGR526 is no longer capable of inducing the activation of the gene preceded by GRE. Tyedmers et al. (2000) checked the conditions that trigger the prionic conformation and they found that heat shock is a significantly relevant factor. The cells in which the prionic conformation is induced, the process is promoted in an autocatalytic manner and all the protein is found in the prionic conformation. The cells resulting show the phenotype [PSI+].
It is important to note that the rate of the change of conformation is not equal to zero even at optimal growth conditions, and that not all the cells become [PSI+]. The rate of spontaneous activation of the switch is thought to be around 10-6 or 10-7 (Alberti et al., 2009). This process is promoted under heat stress (Tyedmers et al., 2008), probably because of the important role of heat shock proteins like Hsp104 in the formation and maintenance of the amyloid fiber (Halfmann et al., 2009). These approximate rates will have very important implications for the yeastworld model that we briefly describe in the following subsection, and with more detail in our wiki [http://2010.igem.org/Team:Valencia/Modeling Modeling] section.
Yeastworld
A model population of yeast containing the prionic switch growing under low temperatures would contain a pool of [psi -] (black) cells and a minimal amount of [PSI+] (white) cells. The high proportion of black yeasts will cause an increase of the planet mean temperature (as can be deduced by the Microbial Albedo Recorder section of the project). As we explained in the section above, when the temperature is high enough, the prionic switch will tend to become activated. Then, there will be a much higher proportion of non-melanic [PSI+] cells. In addition to the underlying molecular tendency, the black cells will be selected against in this situation, because of the increasing of their inner temperature, in comparison to the white cells, that will provoke a reduction of their growth rate. The white cells, on the other hand, have an intrinsic protection against overheating due to their higher albedo. Thus, these cells will grow faster and become more and more abundant. The eventual high proportion of white cells will lead to an increase of the planetary albedo effect and a decrease of the mean temperature. This situation will become autoregulated and reach an equilibrium through several oscillatory cycles. The background and the dynamics of this process can be regarded with much more detail in the [http://2010.igem.org/Team:Valencia/Modeling Modeling] section of the project, but the conclusion is quite remarkable: a pleasant equilibrium temperature of about 30ºC.
Results
We succeeded to transform the yeast strain 5523 with the two plasmids: PG1-NMGR526 and pL2/GZ. In the Fig.3 we demonstrate the obtaining of this double transformation, where it can be seen that this strain was the only one that could grow on a selective medium, compared to others that did not possessed both plasmids.
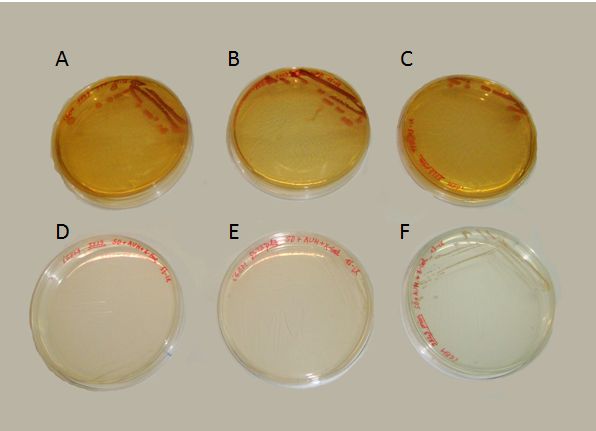
Where growing in presence of X-gal, the proper substrate for the beta-galactosidase enzyme (our reporter gene for the behaviour of the prionic switch), we obtained both blue and white colonies. These colonies can be identified as the [psi-] and [PSI+] phenotypes. In contrast with the results of Li and Lindquist (2000) or Tyedmers et al. (2008), the proportion of white yeasts growing in our plates was unexpectedly high. Further optimization of the experimental manipulation would be needed for the proper characterization of the system.
Nevertheless, the results of the presence of both blue and white colonies and its temperature diference,would be sufficient to recall the properties of the system at the population level.
In order to carachterize the temperature difference between the white and blue colonies we conducted an experiment with our [http://2010.igem.org/Team:Valencia/WB Microbial Albedo Recorder].
Then, the Yeastworld model (described above and, in more detail, in the [http://2010.igem.org/Team:Valencia/Modeling Modeling] section) could depict the evolution and fate of a population of yeast containing the prionic system.
Original source
The chimeric NMGR526 protein was originally constructed by Li and Lindquist (2000). We have to thank Dr. Susan Lindquist (Whitehead Institute for Biomedical Research / Howard Hughes Medical Institute / Department of Biology, MIT) for kindly sending us the prionic system transformed in E.coli strains.
Improvements
The 2015 [http://2015.igem.org/Team:Kent Kent] iGEM team improved this BioBrick by removing all illegal cutting sites within the coding region to improve compatibility with the BioBrick standard. . Kent produced Sup35NM BioBricks, one containing only Sup35 (Part name: BBa_K1739000) and one containing both Sup35NM and the CsgAss signal sequence that targets the protein for export (Part name: BBa_K1739002).
References
- Alberti, S., Halfmann, R., King, O., Kapila, A., Lindquist, S. (2009). A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 137: 146-158.
- Cobb, N.J., Surewicz, W. K. (2009). Prion diseases and their biochemical mechanisms. Biochemistry. 48: 2574–2585.
- Cox,B.S. (1965). A cytoplasmic suppressor of super-suppressor in yeast. Heredity. 20: 505–521.
- Halfmann, R., Alberti, S., Lindquist, S. (2009). Prions, protein homeostasis and phenotypic diversity. Trends in Cell Biology. 20: 125-133.
- Heitzer, M.D., Wolf, I.M., Sanchez, E.R., Witchel, S.F., DeFranco, D.B. (2007). Glucocorticoid receptor phisiology. Rev. Endocr. Metab. Disord.. 8: 321-330.
- Li, L., Lindquist, S. (2000). Creating a protein-based element of inheritance. Science. 287: 661-664.
- Prusiner, S.B. (1982). Novel proteinaceous infectious particles cause scrapie. Science. 216: 136–144.
- Prusiner, S.B. (1998). Prions. PNAS. 95: 13363–13383.
- Si, K., Lindquist, S. Kandel, E.R. (2003). A neuronal isoform of the Aplysia CPEB has prion-like properties. Cell. 115: 879-891.
- Schena, M., Yamamoto, K.R. (1988). Mammalian glucocorticoid receptor derivatives enhance transcription in yeast. Science. 241: 965-967.
- Shorter, J., Lindquist, S. (2005). Prions as adaptive conduits of memory and inheritance. Nature. 6: 435-450.
- True, H.L., Lindquist, S. (2000). A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 407: 477-483.
- Tyedmers, J., Madariaga, M.L., Lindquist, S. (2008). Prion switching in response to environmental stress. PLoS Biology. 6: e294.
- Wickner, R.B. (1994). [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 264: 566-569.
- Wickner, R.B., Shewmaker, F., Kryndushkin, D., Edskes, H.K. (2008). Protein inheritance (prions) based on parallel in-register β-sheet amyloid structures. BioEssays. 30: 955-964.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 122
Illegal PstI site found at 2323 - 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 122
Illegal PstI site found at 2323 - 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 122
Illegal PstI site found at 2323 - 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 122
Illegal PstI site found at 2323 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 2009