Difference between revisions of "Part:BBa K1789015"
Sunjiaqi13 (Talk | contribs) |
Fandongyu13 (Talk | contribs) |
||
| (11 intermediate revisions by 3 users not shown) | |||
| Line 4: | Line 4: | ||
This device is a negative control of GFP with scaf. | This device is a negative control of GFP with scaf. | ||
| − | + | ==Usage and Biology== | |
| − | + | This is the nagative GFP. Using the DNA-binding characteristic of the TAL effector, we can generate a new method to increase the production of | |
| + | heterogenous multi-enzymatic reactions in Prokaryotic cells by rationally designed TALE proteins fused with specific | ||
| + | |||
| + | enzymes and their corresponding DNA sequences (as known as Binding Motifs [BMs]). Here we used Escherichia coli as our | ||
| + | |||
| + | chassis. A plasmid backbone was used as the scaffold for those BMs. The same plasmid was used to encode the fusion protein. | ||
| + | |||
| + | Thus, enzymes fused with TALE proteins could be gathered around the DNA scaffolds, enrich the local enzyme concentration, | ||
| + | |||
| + | and promote the rate of reaction. | ||
| + | |||
| + | This is the first experimental group of our project. Our theory is feasible if the functional parameter of this group is | ||
| + | |||
| + | stronger than the group of negative control. | ||
| + | |||
| + | Split GFP is a technique that has been widely used in the research of protein-protein interaction. In our project, we | ||
| + | |||
| + | demonstrated a prototype by fusing the Amino (or Carboxyl) Half of GFP with TALE1 (or TALE2/3). | ||
| + | |||
| + | By integrating the coding sequences of the TALE-fused proteins and the scaffold, three different plasmids can be | ||
| + | |||
| + | constructed and this is the second one. | ||
| + | |||
| + | ==Sequence and Features== | ||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K1789015 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1789015 SequenceAndFeatures</partinfo> | ||
| + | ==Experimental Validation== | ||
| + | |||
| + | This part is validated through four ways: Enzyme cutting, PCR, and Sequence | ||
| + | |||
| + | ===PCR=== | ||
| + | |||
| + | '''Methods''' | ||
| + | |||
| + | The PCR is performed with Premix EX Taq by Takara. | ||
| + | |||
| + | The PCR protocol is selected based on the Users Manuel. | ||
| + | The Electrophoresis was performed on a 1% Agarose glu. | ||
| + | |||
| + | '''Results''' | ||
| + | |||
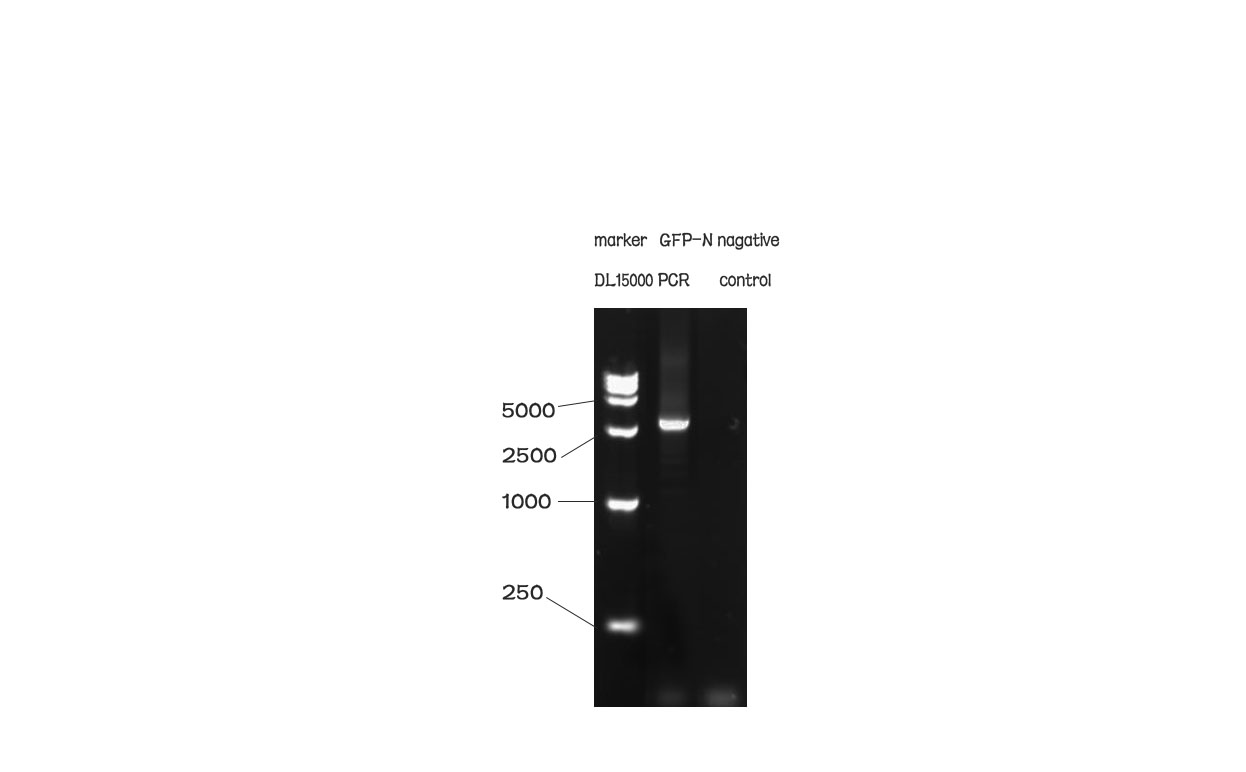
| + | [[File:N1PCR.jpg|500px|]] | ||
| + | |||
| + | Fig. 1 The result of the agarose electrophoresis was shown on the picture above. | ||
| + | |||
| + | ===Enzyme cutting=== | ||
| + | |||
| + | '''Methods''' | ||
| + | |||
| + | After the assembly ,the plasmid was transferred into the Competent ''E. coli'' DH5α). After culturing overnight in LB,we minipreped the plasmid for cutting. | ||
| + | The preparation of the plasmid was performed with TIANprep Mini Plasmid Kit from ''TIANGEN''. The cutting procedure was performed with EcoRI and XbaI restriction endonuclease bought from ''TAKARA''. | ||
| + | |||
| + | The plasmid was cutted in a 20μL system at 37 ℃ for 2 hours. | ||
| + | The Electrophoresis was performed on a 1% Agarose glu. | ||
| + | |||
| + | '''Results''' | ||
| + | |||
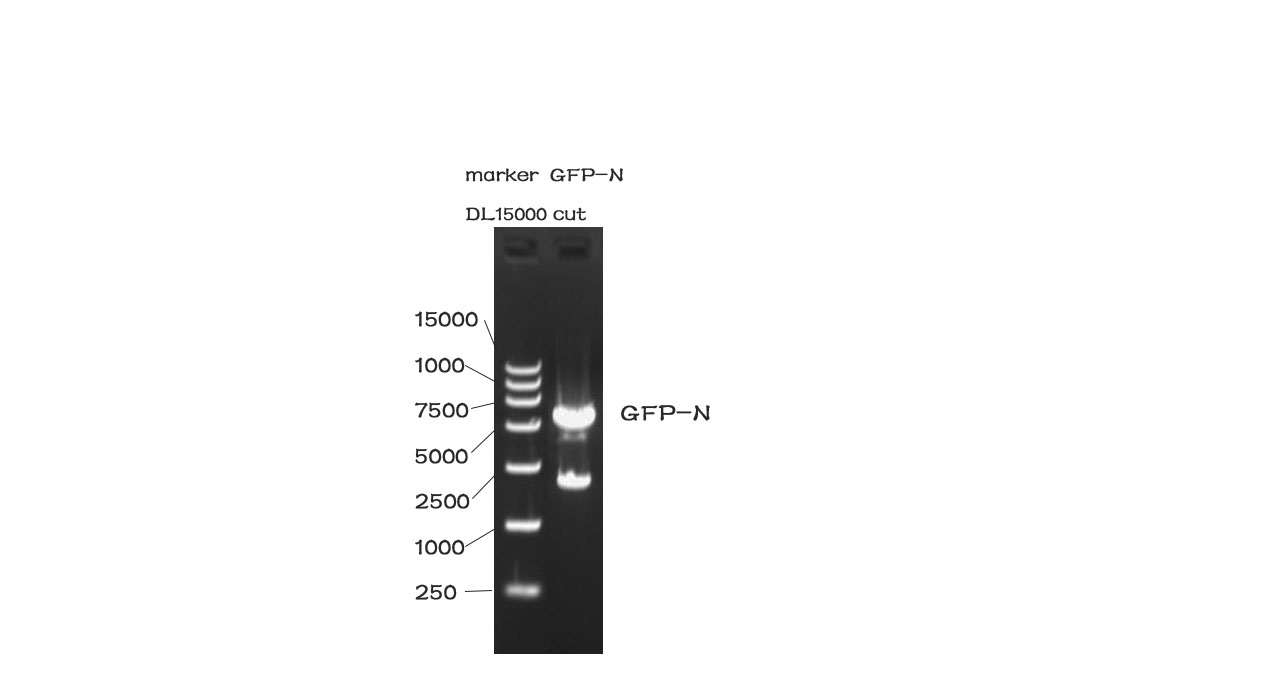
| + | [[File:N1.jpg|500px|]] | ||
| + | |||
| + | Fig. 2The result of the agarose electrophoresis was shown on the picture above. | ||
| − | |||
===Functional Parameters=== | ===Functional Parameters=== | ||
<partinfo>BBa_K1789015 parameters</partinfo> | <partinfo>BBa_K1789015 parameters</partinfo> | ||
| Line 19: | Line 75: | ||
Functional check: | Functional check: | ||
| − | + | Methodology: In order to investigate whether the fluorescence | |
| − | + | observed was due to the presence of the original GFP-N2 construction. | |
| − | + | First of all, the recombinant colonies were transferred into | |
| + | LB culture medium with chloramphenicol, and shook overnight at 37 ℃, | ||
| + | then took the medium for PCR templet. Meanwhile, we extracted plasmid DNA | ||
| + | using a miniprep kit and did a digest with Xbal1 and Pst1 to check | ||
| + | for the presence of the original insert and size of the unfolded vector, respectively. | ||
| + | Result and discussion: The DNA positive strand and the insert | ||
| + | is very clearly visible at just below 3kb and 5kb. | ||
| + | This proves the presence of GFP-N2 in culture. | ||
Latest revision as of 02:38, 19 September 2015
GFP_N1
This device is a negative control of GFP with scaf.
Usage and Biology
This is the nagative GFP. Using the DNA-binding characteristic of the TAL effector, we can generate a new method to increase the production of
heterogenous multi-enzymatic reactions in Prokaryotic cells by rationally designed TALE proteins fused with specific
enzymes and their corresponding DNA sequences (as known as Binding Motifs [BMs]). Here we used Escherichia coli as our
chassis. A plasmid backbone was used as the scaffold for those BMs. The same plasmid was used to encode the fusion protein.
Thus, enzymes fused with TALE proteins could be gathered around the DNA scaffolds, enrich the local enzyme concentration,
and promote the rate of reaction.
This is the first experimental group of our project. Our theory is feasible if the functional parameter of this group is
stronger than the group of negative control.
Split GFP is a technique that has been widely used in the research of protein-protein interaction. In our project, we
demonstrated a prototype by fusing the Amino (or Carboxyl) Half of GFP with TALE1 (or TALE2/3).
By integrating the coding sequences of the TALE-fused proteins and the scaffold, three different plasmids can be
constructed and this is the second one.
Sequence and Features
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 2311
Illegal BamHI site found at 4908 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1304
Illegal BsaI.rc site found at 3390
Illegal BsaI.rc site found at 3798
Illegal BsaI.rc site found at 4104
Illegal BsaI.rc site found at 5094
Experimental Validation
This part is validated through four ways: Enzyme cutting, PCR, and Sequence
PCR
Methods
The PCR is performed with Premix EX Taq by Takara.
The PCR protocol is selected based on the Users Manuel. The Electrophoresis was performed on a 1% Agarose glu.
Results
Fig. 1 The result of the agarose electrophoresis was shown on the picture above.
Enzyme cutting
Methods
After the assembly ,the plasmid was transferred into the Competent E. coli DH5α). After culturing overnight in LB,we minipreped the plasmid for cutting. The preparation of the plasmid was performed with TIANprep Mini Plasmid Kit from TIANGEN. The cutting procedure was performed with EcoRI and XbaI restriction endonuclease bought from TAKARA.
The plasmid was cutted in a 20μL system at 37 ℃ for 2 hours. The Electrophoresis was performed on a 1% Agarose glu.
Results
Fig. 2The result of the agarose electrophoresis was shown on the picture above.
Functional Parameters
Functional check:
Methodology: In order to investigate whether the fluorescence
observed was due to the presence of the original GFP-N2 construction.
First of all, the recombinant colonies were transferred into
LB culture medium with chloramphenicol, and shook overnight at 37 ℃,
then took the medium for PCR templet. Meanwhile, we extracted plasmid DNA
using a miniprep kit and did a digest with Xbal1 and Pst1 to check
for the presence of the original insert and size of the unfolded vector, respectively.
Result and discussion: The DNA positive strand and the insert
is very clearly visible at just below 3kb and 5kb.
This proves the presence of GFP-N2 in culture.