Difference between revisions of "Part:BBa K1824000"
(→Characterization) |
|||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 11: | Line 11: | ||
[[Image:A1-J23119.png|600px]] | [[Image:A1-J23119.png|600px]] | ||
<br>'''Figure 2:''' The fluorescence of J23119+A1 with different dilution ratio at 30℃ and 37℃. | <br>'''Figure 2:''' The fluorescence of J23119+A1 with different dilution ratio at 30℃ and 37℃. | ||
| − | As Figure 2 shows, the fluorescence of the sample at 37℃ is 12 times higher than the fluorescence of the sample at 30℃, which means the amount of protein express at 37℃ is 12 times bigger than at 30℃. | + | <br> |
| + | <br> | ||
| + | <br>eGFP protein was used as report protein to show the function of ribothermometer. The bacteria were centrifuged and observed under UV light by eyes directly. Then the bacteria were sonicated and plate reader was used to measure the fluorescence of eGFP. | ||
| + | <br>As Figure 2 shows, the fluorescence of the sample at 37℃ is 12 times higher than the fluorescence of the sample at 30℃, which means the amount of protein express at 37℃ is 12 times bigger than at 30℃. | ||
| + | |||
| + | ===Measurement and improvement=== | ||
| + | XJTLU-CHINA 2016 iGEM team build a mutagenesis library of A1 using degenerated primers. One set of degenerated primers were designed to mutate the poly-A-loop structure of A1 in order to tight the hairpin structure and the other set of primers were designed to modify Anti-SD of the ribothermometers. The primer sequences used were: | ||
| + | <br> | ||
| + | <p>5'-ACAGAGAACATACTAGAGNNNNNTAAAAAAAAAAAGTACTAAGGAGTACTAG-3'</p> | ||
| + | <br> | ||
| + | <p>5'-AGAGAACATACTAGAGCTCNNNNNNNNNNNNNNGTACTAAGGAGTACTAG-3'</p> | ||
| + | <br> | ||
| + | <p>The mutagenesis pool of A1 was downstream linked to an mRFP reporter gene (see Fig.2) in order to determine the protein expression efficiently. Total 96 colonies were picked after the transformation of mutagenesis library and initially screened using 96 wells plates at 26 ℃ and 37 ℃. Ten colonies with the viable difference in color were selected for further analysis.</p> | ||
| + | |||
| + | <p>[[File:RFP Expressing under control of ribothermometer.jpg|500px|thumb|left|Figure 2. The mRFP expression construct that used for the screening of effective ribothermometers]]</p> | ||
| + | |||
| + | <p> | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | These colonies were seeded in 150ml flasks that contain 20ml LB culture for 12 hrs. The fluorescent was then measured at an excited<br>length of 580nm and emitted length of 607nm. OD 600 of each sample was measured at the same time.</p> | ||
| + | <br>[[File:Eb rfp.JPG|500px|thumb|left|Figure 3. The difference in performance of sensing temperature of 10 A1 mutants.]] | ||
| + | <br><br><br><br><br><br><br><br><br> | ||
| + | <br><br><br><br><br><br><br><br><br><br> | ||
| + | All 10 obtained mutants were sent for sequencing and the result is included in the table below. | ||
| + | |||
| + | [[File:Updated sequencing table RNAT .jpg|500px|thumb|left|Table 1. Sequencing results of 10 mutants with broad-spectrum of sensitivity in respond to temperature]] | ||
===References=== | ===References=== | ||
| Line 26: | Line 50: | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
| + | |||
| + | ===Characterlization and improvement === | ||
| + | XJTLU-CHINA 2016 iGEM team build a mutagenesis library of A1 using degenerated primers. One set of degenerated primers were designed to mutate the poly-A-loop structure of A1 in order to tight the hairpin structure and the other set of primers were designed to modify Anti-SD of the ribothermometers. The primer sequences used were: | ||
| + | <br> | ||
| + | <p>5'-ACAGAGAACATACTAGAGNNNNNTAAAAAAAAAAAGTACTAAGGAGTACTAG-3'</p> | ||
| + | <br> | ||
| + | <p>5'-AGAGAACATACTAGAGCTCNNNNNNNNNNNNNNGTACTAAGGAGTACTAG-3'</p> | ||
| + | <br> | ||
| + | <p>The mutagenesis pool of A1 was downstream linked to an mRFP reporter gene (see Fig.2) in order to determine the protein expression efficiently. Total 96 colonies were picked after the transformation of mutagenesis library and initially screened using 96 wells plates at 26 ℃ and 37 ℃. Ten colonies with the viable difference in color were selected for further analysis.</p> | ||
| + | |||
| + | <p>[[File:RFP Expressing under control of ribothermometer.jpg|500px|thumb|left|Figure 2. The mRFP expression construct that used for the screening of effective ribothermometers]]</p> | ||
| + | |||
| + | <p> | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | These colonies were seeded in 150ml flasks that contain 20ml LB culture for 12 hrs. The fluorescent was then measured at an excited<br>length of 580nm and emitted length of 607nm. OD 600 of each sample was measured at the same time.</p> | ||
| + | <br>[[File:Eb rfp.JPG|500px|thumb|left|Figure 3. The difference in performance of sensing temperature of 10 A1 mutants.]] | ||
| + | <br><br><br><br><br><br><br><br><br> | ||
| + | <br><br><br><br><br><br><br><br><br><br> | ||
| + | All 10 obtained mutants were sent for sequencing and the result is included in the table below. | ||
| + | |||
| + | [[File:Updated sequencing table RNAT .jpg|500px|thumb|left|Table 1. Sequencing results of 10 mutants with broad-spectrum of sensitivity in respond to temperature]] | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
<partinfo>BBa_K1824000 parameters</partinfo> | <partinfo>BBa_K1824000 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
Latest revision as of 16:07, 20 October 2016
Constitutive Promoter J23119 + RNA Thermometer A1
This is a composite regulatory part that consisting of constitutive promoter BBa_J23119 and RNA Thermometer A1 BBa_K1824556 for temperature induced gene expression at 42 degrees. The possible secondary structure of A1 was simulated by RNAstructure (Fig.1). This combination is compatible with the assembly method that XJTLU-CHINA promoted.
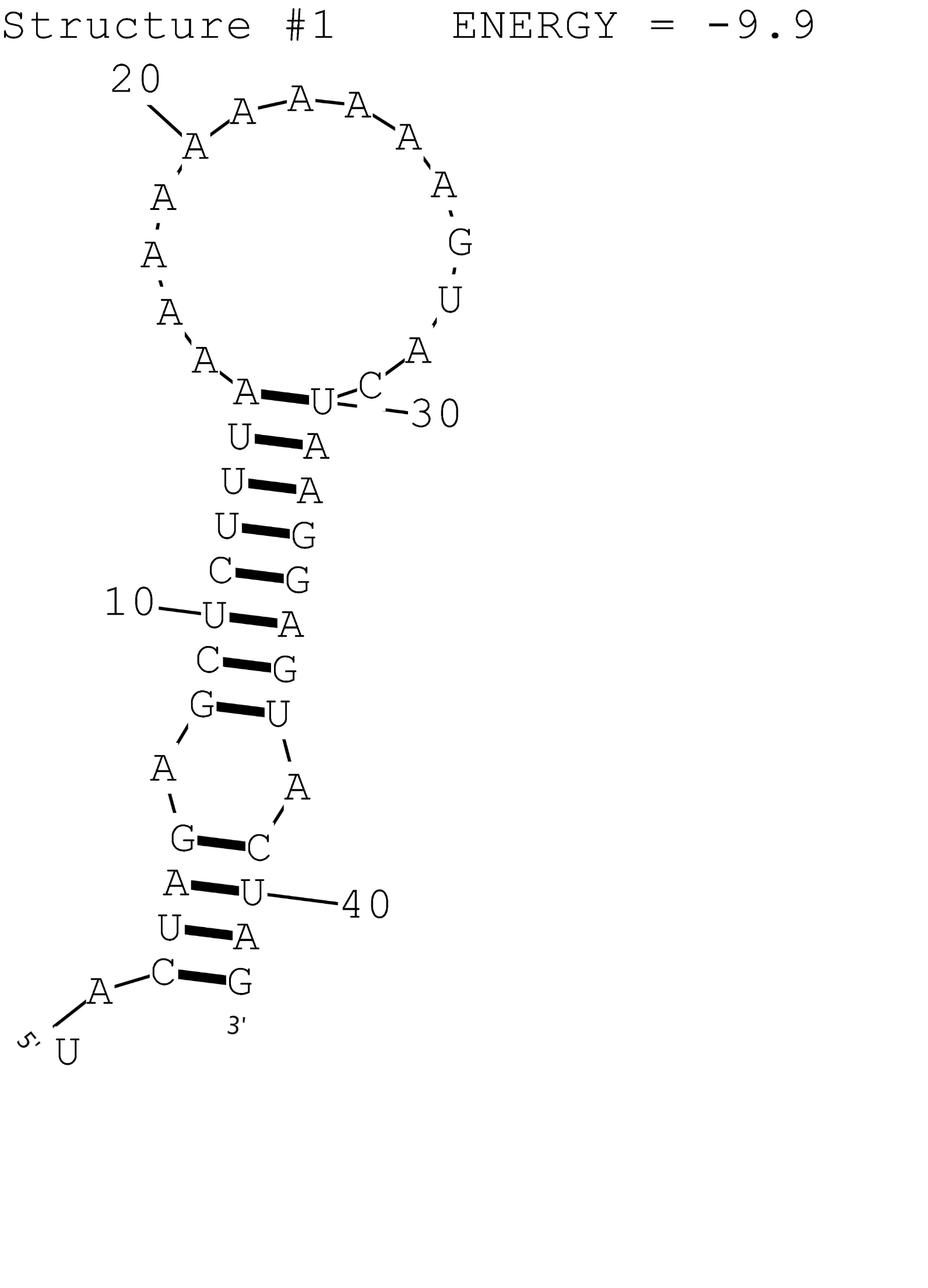
Figure 1: Possible secondary structure of RNAT A1.
Characterization
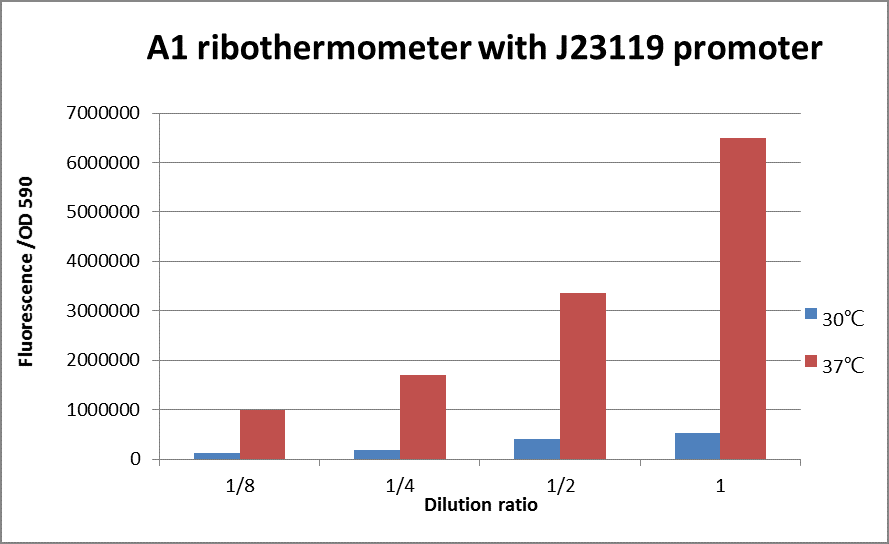
Figure 2: The fluorescence of J23119+A1 with different dilution ratio at 30℃ and 37℃.
eGFP protein was used as report protein to show the function of ribothermometer. The bacteria were centrifuged and observed under UV light by eyes directly. Then the bacteria were sonicated and plate reader was used to measure the fluorescence of eGFP.
As Figure 2 shows, the fluorescence of the sample at 37℃ is 12 times higher than the fluorescence of the sample at 30℃, which means the amount of protein express at 37℃ is 12 times bigger than at 30℃.
Measurement and improvement
XJTLU-CHINA 2016 iGEM team build a mutagenesis library of A1 using degenerated primers. One set of degenerated primers were designed to mutate the poly-A-loop structure of A1 in order to tight the hairpin structure and the other set of primers were designed to modify Anti-SD of the ribothermometers. The primer sequences used were:
5'-ACAGAGAACATACTAGAGNNNNNTAAAAAAAAAAAGTACTAAGGAGTACTAG-3'
5'-AGAGAACATACTAGAGCTCNNNNNNNNNNNNNNGTACTAAGGAGTACTAG-3'
The mutagenesis pool of A1 was downstream linked to an mRFP reporter gene (see Fig.2) in order to determine the protein expression efficiently. Total 96 colonies were picked after the transformation of mutagenesis library and initially screened using 96 wells plates at 26 ℃ and 37 ℃. Ten colonies with the viable difference in color were selected for further analysis.
These colonies were seeded in 150ml flasks that contain 20ml LB culture for 12 hrs. The fluorescent was then measured at an excited
length of 580nm and emitted length of 607nm. OD 600 of each sample was measured at the same time.
All 10 obtained mutants were sent for sequencing and the result is included in the table below.
References
1. Nocker, A. et al. mRNA-based thermosensor controls expression of rhizobial heat shock genes. Nucleic Acids Res. 29, 4800–4807 (2001).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]



