Difference between revisions of "Part:BBa K1696003"
Ggjyliuyue (Talk | contribs) |
|||
| (7 intermediate revisions by 3 users not shown) | |||
| Line 15: | Line 15: | ||
<html> | <html> | ||
| − | <img src=" https://static.igem.org/mediawiki/parts/2/ | + | <img src=" https://static.igem.org/mediawiki/parts/2/2b/LdhE-3.png" width="500px"/> |
</html> | </html> | ||
| − | Fig2. The lactate yield of BL21 and BL21+ldhE. | + | Fig2. The lactate yield of BL21 and BL21+ldhE in flask-shaking fermentation condition (LB medium). |
| + | |||
| + | '''pflB knockout''' | ||
| + | |||
| + | In order to enhance the transformation efficiency from glucose to lactate, as well as improve the characterization of our ldhE part, we decide to knockout the the pflB and poxB. | ||
| + | |||
| + | <html> | ||
| + | <img src=" https://static.igem.org/mediawiki/2015/1/19/LdhE.png" width="300px"/> | ||
| + | </html> | ||
| + | |||
| + | Figure 3. Lactate metabolic pathway. The pflB and poxB knocking out and ldhE insertion will result in the accumulation of lactate. | ||
| + | |||
| + | |||
| + | PFL is a crucial enzyme in the glucose metabolism under anaerobic condition, which can catalyze one molecule of pyruvate to one molecule of formicate and one molecule of acetylcoenzyme A (AcCoA). According to metabolic pathway, pyruvate is consumed both in the PFL and LDH reactions. In the wild type E. coli, LDH reaction is not as competitive as the reaction through PFL, and therefore, knockout of the PFL related genes will contribute to redistribute the metabolic flux. When the PFL pathway is blocked, E. coli will conceivably alter the distribution of these products to overcome the imbalanced reducing equivalents caused by the pathway knockout. Therefore, knockout of pflB not only decreased the carbon flow to acetyl-CoA under anaerobic condition but also reduced the anaerobic consumption of NADH through reductive TCA. | ||
| + | |||
| + | After the knockout of pflB, though the growth rate was slightly slowed down, the yield of lactate of engineered strain was improved greatly and showed a significant difference from the wild type MG1655. From the result, we can see our part functioned as expected. | ||
| + | |||
| + | <html> | ||
| + | <img src=" https://static.igem.org/mediawiki/2015/a/a6/屏幕快照_2015-09-16_下午11.16.03.png" width="700px"/> | ||
| + | </html> | ||
| + | |||
| + | Fig4.Comparison diagram of lactate production of three different strains. MG1655: wild type ; MG1655ΔpflB: pflB knockout, ldhE blank; MG1655ΔpflB+ldhE: containing the functional ldhE part we design | ||
| + | |||
| + | '''Knockout strategy''' | ||
| + | |||
| + | For genes knockout, we adopted a effective, easy-to-use two-step system in which the cell is first transformed with a helper plasmid harboring genes encoding the λ-Red enzymes, I-SceI endonuclease, and RecA. λ-Red enzymes expressed from the helper plasmid are used to recombineer a small ‘landing pad’, a tetracycline resistance gene (tetA) flanked by I-SceI recognition sites and landing pad regions, into the desired location in the chromosome. After tetracycline selection for successful landing pad integrants, the cell is transformed with a donor plasmid carrying the desired insertion fragment; this fragment is excised by I-SceI and incorporated into the landing pad via recombination at the landing pad regions. | ||
| + | |||
| + | First round of recombineering consists of three rounds of PCR for the construction of recombinant genome cassettes which subsequently positively selected by selection markers such as Tetr. In the second step, TetA marker was released by simultaneous induction of I-SceI and Red recombinase expression which serves as a negative selection. | ||
| + | |||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/2015/4/48/Lambda_red.png" width="700px"/> | ||
| + | </html> | ||
| + | |||
| + | Fig5. First step in the new two-step scar-less gene deletion using λ-Red recombineering method. Three rounds of PCR together with tet selection make up of our recombinant. | ||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 32: | Line 66: | ||
<partinfo>BBa_K1696003 parameters</partinfo> | <partinfo>BBa_K1696003 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | <html> | ||
| + | <style> | ||
| + | .markdown img { | ||
| + | display: block; | ||
| + | padding: 5px; | ||
| + | margin: 0 auto; | ||
| + | max-width: 100%; | ||
| + | height: auto; | ||
| + | } | ||
| + | .markdown .img-gallery { | ||
| + | display: flex; | ||
| + | flex-wrap: wrap; | ||
| + | align-items: center; | ||
| + | align-content: center; | ||
| + | } | ||
| + | .markdown img + em { | ||
| + | font-style: italic; | ||
| + | display: inherit; | ||
| + | text-align: center; | ||
| + | font-size: 90%; | ||
| + | } | ||
| + | .markdown .img-gallery > span { | ||
| + | margin: 0 auto; | ||
| + | } | ||
| + | .markdown h1, | ||
| + | .markdown h2 { | ||
| + | padding-top: 65px; | ||
| + | margin-top: -65px; | ||
| + | } | ||
| + | .markdown h1 { | ||
| + | color: #155; | ||
| + | } | ||
| + | .markdown h2 { | ||
| + | color: #377; | ||
| + | } | ||
| + | .markdown p { | ||
| + | color: #333; | ||
| + | } | ||
| + | .markdown video { | ||
| + | width: 100%; | ||
| + | margin-left: auto; | ||
| + | margin-right: auto; | ||
| + | } | ||
| + | </style> | ||
| + | <div class="markdown"> | ||
| + | <h1>Optimization of the characterization</h1> | ||
| + | <p>This BioBrick codes for <em>ldhA </em>gene from <em>Lactobacillus </em>and allows a higher production and secretion of lactate. This gene, which can also be found in <em>E. coli.</em></p> | ||
| + | <p>To optimize it characterization, we performed a PCR with primers which as some specificity with the sequence and ran the PCR product on an agarose gel. This proves the presence of the gene in the BioBrick. Furthermore, we recovered the sequence, inserted it in one of our backbones, and transformed a K12 <em>E. coli </em>bacterium. We then tested its efficacy by measuring pH (it should be more acidic as lactate accumulate).</p> | ||
| + | <h1>Gel migration of PCR products</h1> | ||
| + | <p><img src="https://static.igem.org/mediawiki/parts/a/a9/T--Ionis_Paris--ldha_1.jpg" alt="Agarose gel photo" /> | ||
| + | <em>Figure 1: 1% agarose gel of ldhA</em></p> | ||
| + | <p>The part has a size of 1158 bp and our ldhA (BBa_K3102006) has a size of 1053 pb. Two strips are observed at the correct sizes. The ldhA strip seems more fainted which can be explained by a non 100% specificity of our primers.</p> | ||
| + | <p>Those results confirmed the presence of the <em>ldhA </em>gene in the BioBrick.</p> | ||
| + | <h1>Determination of <em>ldhA </em>efficiency via pH measurements</h1> | ||
| + | <p><img src="https://static.igem.org/mediawiki/parts/e/e0/T--Ionis_Paris--ldha_image1.png" alt="pH evolution chart" /> | ||
| + | <em>Figure 2: a. Evolution of pH in ldhA and control samples b. Evolution of pH ratio c. Measurement of pH using indicator strips, t=0h d. Measurement of pH using indicator strips, t=20h e.pH scale</em></p> | ||
| + | <p>From these results, two tendencies are observed: control bacteria culture seems to become more basic overtime (from pH 7.0 to pH 8.5) whereas the <em>ldhA </em>transformed culture tends to get more acidic (from pH 7 to pH 5). This indicates the efficiency of the <em>ldhA </em>gene as the acidity measured must be due to acetate production and secretion, counterbalancing the basic molecules produced.</p> | ||
| + | </div> | ||
| + | </html> | ||
Latest revision as of 08:47, 21 October 2019
L-lactate producing part1
L-Lactic acid, one of the most important chiral molecules and organic acids, is produced via pyruvate from carbohydrates in diverse microorganisms catalyzed by an NAD+-dependent L-lactate dehydrogenase (ldhA).[1]LDH is an enzyme found in nearly all living cells (animals, plants, and prokaryotes), which catalyzes the conversion of pyruvate to lactate with NADH serving as the coenzyme. In our project, we intend to introduce high-yield exogenous L-(+)-lactate dehydrogenase gene (ldhA) from Lactobacillus, which can produce a relatively larger amount of L-lactate. Similar metabolic pathway has been found in a variety of organisms, to distinguish the differences, we renamed the heterogenous L-lactate dehydrogenase ldhE while homogenous one keeps original.
We choose a strong promoter+ strong RBS to construct L-Lactate part. The NAD+-dependent L-lactate dehydrogenase gene is from Lactobacillus casei.

Fig.1 Design of the part.
But we found the yield was no different between BL21(blank) and BL21+ldhE(experientmental). Shown in Fig2.

Fig2. The lactate yield of BL21 and BL21+ldhE in flask-shaking fermentation condition (LB medium).
pflB knockout
In order to enhance the transformation efficiency from glucose to lactate, as well as improve the characterization of our ldhE part, we decide to knockout the the pflB and poxB.

Figure 3. Lactate metabolic pathway. The pflB and poxB knocking out and ldhE insertion will result in the accumulation of lactate.
PFL is a crucial enzyme in the glucose metabolism under anaerobic condition, which can catalyze one molecule of pyruvate to one molecule of formicate and one molecule of acetylcoenzyme A (AcCoA). According to metabolic pathway, pyruvate is consumed both in the PFL and LDH reactions. In the wild type E. coli, LDH reaction is not as competitive as the reaction through PFL, and therefore, knockout of the PFL related genes will contribute to redistribute the metabolic flux. When the PFL pathway is blocked, E. coli will conceivably alter the distribution of these products to overcome the imbalanced reducing equivalents caused by the pathway knockout. Therefore, knockout of pflB not only decreased the carbon flow to acetyl-CoA under anaerobic condition but also reduced the anaerobic consumption of NADH through reductive TCA.
After the knockout of pflB, though the growth rate was slightly slowed down, the yield of lactate of engineered strain was improved greatly and showed a significant difference from the wild type MG1655. From the result, we can see our part functioned as expected.
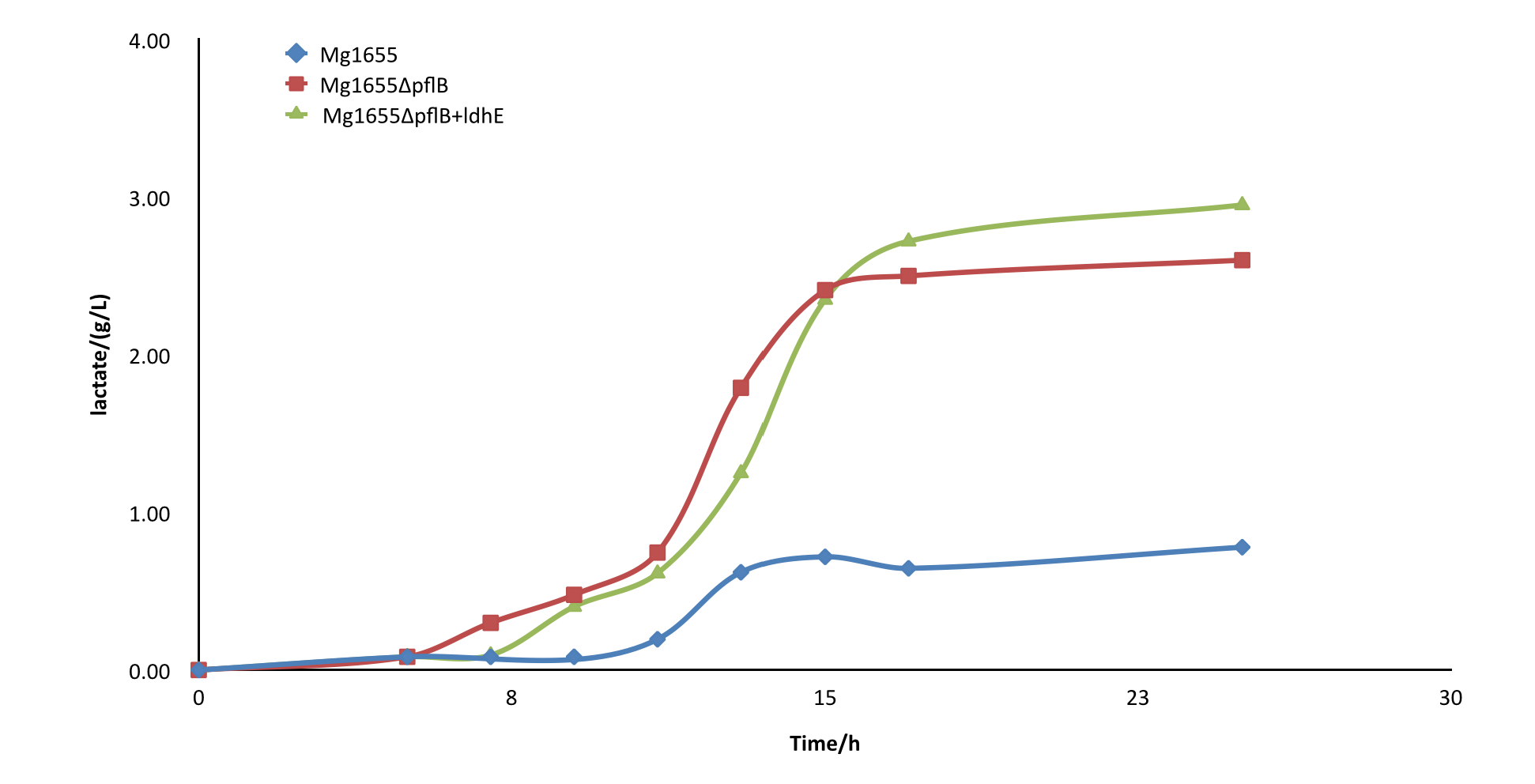
Fig4.Comparison diagram of lactate production of three different strains. MG1655: wild type ; MG1655ΔpflB: pflB knockout, ldhE blank; MG1655ΔpflB+ldhE: containing the functional ldhE part we design
Knockout strategy
For genes knockout, we adopted a effective, easy-to-use two-step system in which the cell is first transformed with a helper plasmid harboring genes encoding the λ-Red enzymes, I-SceI endonuclease, and RecA. λ-Red enzymes expressed from the helper plasmid are used to recombineer a small ‘landing pad’, a tetracycline resistance gene (tetA) flanked by I-SceI recognition sites and landing pad regions, into the desired location in the chromosome. After tetracycline selection for successful landing pad integrants, the cell is transformed with a donor plasmid carrying the desired insertion fragment; this fragment is excised by I-SceI and incorporated into the landing pad via recombination at the landing pad regions.
First round of recombineering consists of three rounds of PCR for the construction of recombinant genome cassettes which subsequently positively selected by selection markers such as Tetr. In the second step, TetA marker was released by simultaneous induction of I-SceI and Red recombinase expression which serves as a negative selection.

Fig5. First step in the new two-step scar-less gene deletion using λ-Red recombineering method. Three rounds of PCR together with tet selection make up of our recombinant.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 36
Illegal BsaI.rc site found at 1058
Optimization of the characterization
This BioBrick codes for ldhA gene from Lactobacillus and allows a higher production and secretion of lactate. This gene, which can also be found in E. coli.
To optimize it characterization, we performed a PCR with primers which as some specificity with the sequence and ran the PCR product on an agarose gel. This proves the presence of the gene in the BioBrick. Furthermore, we recovered the sequence, inserted it in one of our backbones, and transformed a K12 E. coli bacterium. We then tested its efficacy by measuring pH (it should be more acidic as lactate accumulate).
Gel migration of PCR products
 Figure 1: 1% agarose gel of ldhA
Figure 1: 1% agarose gel of ldhA
The part has a size of 1158 bp and our ldhA (BBa_K3102006) has a size of 1053 pb. Two strips are observed at the correct sizes. The ldhA strip seems more fainted which can be explained by a non 100% specificity of our primers.
Those results confirmed the presence of the ldhA gene in the BioBrick.
Determination of ldhA efficiency via pH measurements
 Figure 2: a. Evolution of pH in ldhA and control samples b. Evolution of pH ratio c. Measurement of pH using indicator strips, t=0h d. Measurement of pH using indicator strips, t=20h e.pH scale
Figure 2: a. Evolution of pH in ldhA and control samples b. Evolution of pH ratio c. Measurement of pH using indicator strips, t=0h d. Measurement of pH using indicator strips, t=20h e.pH scale
From these results, two tendencies are observed: control bacteria culture seems to become more basic overtime (from pH 7.0 to pH 8.5) whereas the ldhA transformed culture tends to get more acidic (from pH 7 to pH 5). This indicates the efficiency of the ldhA gene as the acidity measured must be due to acetate production and secretion, counterbalancing the basic molecules produced.
