Difference between revisions of "Part:BBa K1720001"
(→Experiment 1:) |
|||
| (10 intermediate revisions by the same user not shown) | |||
| Line 22: | Line 22: | ||
| − | + | In the preliminary experiments, we designed a lentiviral vector containing our part and packaged the vector with lentivirus. The sequence to encode a fluorescent reporter was inserted after the antibiotics-resistance gene,rather than directly after our part, to avoid any potential influences on the function of the part. | |
| Line 33: | Line 33: | ||
===Virus Titer: (2.24±2)×10^8 TU/mL=== | ===Virus Titer: (2.24±2)×10^8 TU/mL=== | ||
| − | + | Functional titer was determined based on q-PCR amplification of a small fragment from the lentiviral vector, WRPE, which was integrated into the genome of transduced 293T cells. | |
| − | + | ||
===Experiment 1:=== | ===Experiment 1:=== | ||
| − | At the beginning of our experiment, we aimed to prove that HEK293 cells can be transfected by our | + | At the beginning of our experiment, we aimed to prove that HEK293 cells can be transfected by our lentivirus. In our vector we inserted EGFP gene as a reporter.Once HEK293 cells were transfected successfully, green fluorescent signal would be expected under fluorescence microscope. |
<b>Protocol:</b> | <b>Protocol:</b> | ||
| + | |||
1. Seed cells to be 40% confluent at a 35mm culture dish. | 1. Seed cells to be 40% confluent at a 35mm culture dish. | ||
2. | 2. | ||
| Line 61: | Line 61: | ||
<b>Result:</b> | <b>Result:</b> | ||
| − | [[File:beta3 transfection.png|400px|thumb|left|]] | + | [[File:beta3 transfection.png|400px|thumb|left|Green fluorescence signal after transfection of alpha3 subunit]] |
| Line 92: | Line 92: | ||
| − | + | ||
| − | At the same time we also transfected | + | The figure indicated that HEK293 cells had been transfected successfully!At the same time we also transfected HEK293 cells with vectors that contain beta3 subunit and we get the same results. |
===Experiment 2:=== | ===Experiment 2:=== | ||
| Line 185: | Line 185: | ||
<b>Result:</b> | <b>Result:</b> | ||
| − | [[File:betaGel2.png|400px|thumb|left|]] | + | [[File:betaGel2.png|400px|thumb|left|Transcription level after transfection with bata3 subunit]] |
| + | [[File:betaCTmean.png|400px|thumb|left|△CT vs GAPDH after transfection of beta3 subunit]] | ||
| Line 225: | Line 226: | ||
| + | ===Experiment 3:=== | ||
| − | + | As we up-regulate the transcriptional level of sGC alpha3 subunit and bata3 subunit, we used sGC Elisa kit to detect the sGC activity to see whether two subunits combine with each other successfully. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | As we up-regulate the transcriptional level of sGC | + | |
| − | with each other successfully. | + | |
| Line 253: | Line 248: | ||
8. Read the Optical Density ( O . D .) at 450 nm using a Microplate Reader. | 8. Read the Optical Density ( O . D .) at 450 nm using a Microplate Reader. | ||
| + | |||
| Line 258: | Line 254: | ||
1.Standard ( S0 → S5 ) concentration was followed by: 0,30,60,120,240,480 U/L | 1.Standard ( S0 → S5 ) concentration was followed by: 0,30,60,120,240,480 U/L | ||
| − | |||
| + | |||
<b>Result:</b> | <b>Result:</b> | ||
| + | [[File:Elisa-alpha+beta.png|400px|thumb|left|sGC activity after transfection of alpha3 subunit and bata3 subunit]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
===Experiment 4:=== | ===Experiment 4:=== | ||
| − | + | We used sodium nitroprusside(SNP) ,a NO donator that activate sGC and up regulate the level of cGMP, as a positive control. | |
| + | <b>Protocol</b> | ||
| − | + | 1. Seed cells to be 90% confluent at 35mm culture dish. | |
| + | 2. Dilute 20ul SNP (0.1ul mol/L) and 20ul cysteine (0.2ul mol/L) in 1960ul DMEM medium containing 10% FBS | ||
| + | |||
| + | 3.Withdraw culture medium from 35mm culture dish | ||
| + | |||
| + | 4.Add the mixture in step 2 to the cells | ||
| + | |||
| + | 5. Incubate for 10min,20min,30min,60min,90min | ||
| + | |||
| + | 6. Use cGMP Elisa kit to detect the cGMP concentration | ||
| + | |||
| + | <b>Note:</b> | ||
| + | |||
| + | 1. We used cysteine to help SNP release NO | ||
| + | |||
| + | 2.Cysteine will cause interference when we use BCA protein assay kit to detect total protein level as an internal reference, so we make two control. One of the controls was treat by the same amount of cysteine, the other was treat with DMEM medium to cut down the background caused by cysteine. | ||
| + | |||
| + | |||
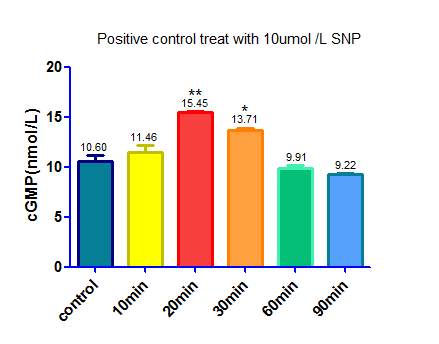
| + | [[File:SCUT China Elisa of SNP.png|400px|thumb|left|cGMP concentration after treated with 10umol/L SNP]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===Experiment 5:=== | ||
| + | After we successfully overexpress sGC, we examined cellular cGMP level using an ELISA kit to examine whether our part can upregulate cGMP levels through sGC overexpression. | ||
| + | |||
| + | |||
| + | <b>protocol:</b> | ||
1. Prepare all standards and samples be added in duplicate to the micro elisa stripplate. | 1. Prepare all standards and samples be added in duplicate to the micro elisa stripplate. | ||
| Line 292: | Line 375: | ||
<b>Result:</b> | <b>Result:</b> | ||
| + | [[File:Elisa-alpha2.png|400px|thumb|left|cGMP concentration after transfection]] | ||
Latest revision as of 02:28, 18 September 2015
Human guanylate cyclase1,soluble, beta 3 unit
This is a part used for coding soluble human guanylate cyclase subunit:beta 3.beta 3 unit interacts with a alpha 3 subunit to form the guanylate cyclase enzyme. Soluble guanylate cyclases are heterodimeric proteins that catalyze the conversion of GTP to 3',5'-cyclic GMP and pyrophosphate. It is important for smooth muscle relaxation in the cardiovascular system. sGC contains an HNOX domain, which serves as a receptor for ligands such as nitric oxide, oxygen and nitrovasodilator drugs.There are several different isoforms of sGC subunit ,but the cardiovascular system abundant with alpha 3 unit and beta 3 unit.
We used lentiviral vector to transfected alpha 3 unit and beta 3 unit to HEK293 cells together and vivo green fluorescence signal was observed under fluorescence microscope, it meant that this part was transfected into HEK293 cells successfully. Then we used qRT-PCR to observe transcriptional level of beta 3 unit. Since soluble guanylate cyclases (sGC) are heterodimeric proteins that catalyze the conversion of GTP to 3',5'-cyclic GMP(cGMP) and pyrophosphate. The level of cGMP will be up regulated.We detected the cGMP concentration with Elisa Kit.The positive control was HEK293 cells that treat with Sodium Nitroprusside ,a NO donator that activate sGC and up regulate the level of cGMP. A negative control was made by transfecting an empty vector that does not contain sGC subunit. We used Elisa Kit to detect cGMP level. The results are as follow:
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 579
Illegal BamHI site found at 800 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
In the preliminary experiments, we designed a lentiviral vector containing our part and packaged the vector with lentivirus. The sequence to encode a fluorescent reporter was inserted after the antibiotics-resistance gene,rather than directly after our part, to avoid any potential influences on the function of the part.
Vector Map:
Vector Components:
Virus Titer: (2.24±2)×10^8 TU/mL
Functional titer was determined based on q-PCR amplification of a small fragment from the lentiviral vector, WRPE, which was integrated into the genome of transduced 293T cells.
Experiment 1:
At the beginning of our experiment, we aimed to prove that HEK293 cells can be transfected by our lentivirus. In our vector we inserted EGFP gene as a reporter.Once HEK293 cells were transfected successfully, green fluorescent signal would be expected under fluorescence microscope.
Protocol:
1. Seed cells to be 40% confluent at a 35mm culture dish. 2. 3. Dilute 10ul lentiviral vector in 1ml DMEM medium containing 10% FBS 4. 3. Withdraw culture medium from 35mm culture dish.
5. Add vector-DMEM complex to cells 6. 5. Incubate for 15 hours.
7. Withdraw vector-DMEM complex from culture dish. 8. 7. Add 2ml DMEM medium containing 10% FBS to cells and incubate for 10 hours
8. Observe the cells under Inverted fluorescence microscope.
Result:
The figure indicated that HEK293 cells had been transfected successfully!At the same time we also transfected HEK293 cells with vectors that contain beta3 subunit and we get the same results.
Experiment 2:
After we proved that HEK293 cells can be transfected, sGC beta3 subunit gene expression levels were determined by PCR.
Protocol:
RNA extraction
1.Add trizol (3ml per culture dish);
2.Keep portions in centrifuge tube(1ml per centrifuge tube)
3.Homogenized by pipetting several times.
4.Incubate samples for 5 min at room temp.
5.Add chloroform (1/5 volume of trizol; e.g. 0.2ml to 1ml)
6.Shake for 15sec.
7.Incubate samples for 5 min at room temp.
8.Centrifuge11.5G, 15 min, 4 ℃.
9.Transfer 0.5ml aqueous phase to a new centrifuge tube.
10.Add isopropanol (1/2 volume of trizol; e.g. 0.5ml to 1ml)
11.Reverse blending.
12.Incubate samples for 10 min at room temp.
13.Centrifuge11.5G, 10 min, 4 ℃.
14.Discard the supernatant.
15.Add 70% EtOH (1 volume of trizol; e.g. 1ml to 1ml ,add & vortex briefly)
16.Centrifuge11.5G, 5 min, 4 ℃.
17.Discard the supernatant.
18.Air-dry pellet for 2-5min.
19.Add 20μlRNase free water and store in -70℃ environment.
20.Determine RNA content by UV spectrophotometry.
21.Electrophoresis of RNA.
Two-Step RT-PCR
STEP1:Reverse Transcription
1.Assemble the reaction on ice. Add the enzyme last.
2.Add the following components to a nuclease-free microcentrifuge tube.

3.Heat mixture to 65°C for 5 min and quick chill on ice. Collect the contents of the tube by brief centrifugation and add:
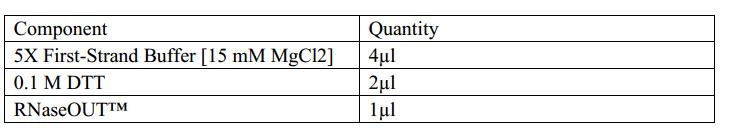
4.Mix contents of the tube gently and incubate at 37°C for 2 min.
5.Add 1 µl (200 units) of M-MLV RT,and mix by pipetting gently up and down.
6.Incubate 50 min at 37°C.
7.Inactivate the reaction by heating at 70°C for 15 min.
STEP2:PCR
1.Add RNase free water 3µl, PowerUp™ SYBR Green Master Mix 5µl, cDNA1µl, Forward primer 0.5µl, Reverse primer 0.5µl each tube in PCR Tubes Strip PCR.
2) Mix the components gently but thoroughly. Then centrifuge brifly to spin down the cetents and eliminate any air bubbles.Incubate in Thermacycler:
a) UDG Activation: 50°C for 2min.
b)AmpliTaq Fast DNA Ploymerase,UP Activation: 95°C for 2min.
c) 40cycles:
Denature: 95°C for 15 sec
Anneal/Extend:60°C for 1min.
Result:
Experiment 3:
As we up-regulate the transcriptional level of sGC alpha3 subunit and bata3 subunit, we used sGC Elisa kit to detect the sGC activity to see whether two subunits combine with each other successfully.
Protocol:
1. Prepare all standards and samples be added in duplicate to the micro elisa stripplate.
2. Add standard : Set Standard wells , testing sample wells. Add standard 50 μl to standard well .
3. Add Sample : Add testing sample 10 μl then add Sample Diluent 40 μl to testing sample well(samples were 5 times diluted )Blank well doesn’t add anyting.
4. Add 100 μl of HRP-conjugate reagent to each well , cover with an adhesive strip and incubate for 60 minutes at 37°C .
5. Aspirate each well and wash by filling each well with Wash Solution (400μl ), repeating the process four times for a total of five washes. After the last wash, remove any remaining Wash Solution by decanting. Invert the plate and blot it against cleanpaper towels.
6. Add chromogen solution A 50μl and chromogen solution B 50μl to each well.Gently mix and incubate for 15 minutes at 37 ℃ Protect from light .
7. Add 50μl Stop Solution to each well.
8. Read the Optical Density ( O . D .) at 450 nm using a Microplate Reader.
Note:
1.Standard ( S0 → S5 ) concentration was followed by: 0,30,60,120,240,480 U/L
Result:
Experiment 4:
We used sodium nitroprusside(SNP) ,a NO donator that activate sGC and up regulate the level of cGMP, as a positive control.
Protocol
1. Seed cells to be 90% confluent at 35mm culture dish.
2. Dilute 20ul SNP (0.1ul mol/L) and 20ul cysteine (0.2ul mol/L) in 1960ul DMEM medium containing 10% FBS
3.Withdraw culture medium from 35mm culture dish
4.Add the mixture in step 2 to the cells
5. Incubate for 10min,20min,30min,60min,90min
6. Use cGMP Elisa kit to detect the cGMP concentration
Note:
1. We used cysteine to help SNP release NO
2.Cysteine will cause interference when we use BCA protein assay kit to detect total protein level as an internal reference, so we make two control. One of the controls was treat by the same amount of cysteine, the other was treat with DMEM medium to cut down the background caused by cysteine.
Experiment 5:
After we successfully overexpress sGC, we examined cellular cGMP level using an ELISA kit to examine whether our part can upregulate cGMP levels through sGC overexpression.
protocol:
1. Prepare all standards and samples be added in duplicate to the micro elisa stripplate.
2. Add standard : Set Standard wells , testing sample wells. Add standard 50 μl to standard well .
3. Add Sample : Add testing sample 10 μl then add Sample Diluent 40 μl to testing sample well (samples were 5 times diluted ) ; Blank well doesn’t add anyting.
4. Add 100 μl of HRP-conjugate reagent to each well , cover with an adhesive strip and incubate for 60 minutes at 37°C .
5. Aspirate each well and wash by filling each well with Wash Solution (400μl ), repeating the process four times for a total of five washes.
6. After the last wash, remove any remaining Wash Solution by decanting. Invert the plate and blot it against clean paper towels.
7. Add chromogen solution A 50μl and chromogen solution B 50μl to each well.Gently mix and incubate for 15 minutes at 37 C . Protect from light .
8. Add 50μl Stop Solution to each well.
9. Read the Optical Density ( O.D ) at 450 nm using a Microplate Reader.
Note:
1.Standard ( S0 → S5 ) concentration was followed by: 0,2,4,8,16,32 nmol/L.
Result: