Difference between revisions of "Part:BBa K1761003"
(adding schematical overviews) |
Pantelistra (Talk | contribs) (→Sequence comparison of mNeonGreen with EGFP) |
||
| (19 intermediate revisions by 5 users not shown) | |||
| Line 4: | Line 4: | ||
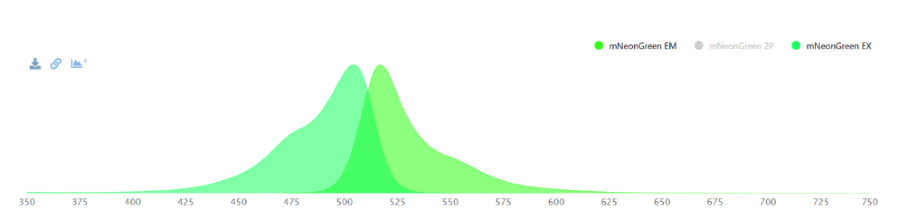
mNeonGreen is a yellow-green fluorescent protein. For a fluorescene measurement, the mNeonGreen fluorophore can be excitated with a laser with a wavelenght of 480 nm and readed out at 517 nm. | mNeonGreen is a yellow-green fluorescent protein. For a fluorescene measurement, the mNeonGreen fluorophore can be excitated with a laser with a wavelenght of 480 nm and readed out at 517 nm. | ||
| + | [[File:Athens2022-mNeonGreen_ex _em.png]] | ||
| + | |||
| + | ''Figure 1: Excitation and Emission wavelengths of mNeonGreen '' | ||
== Usage and Biology == | == Usage and Biology == | ||
| − | mNeonGreen is a yellow-green fluorescent protein and is derived from a tetrameric fluorescent protein from cephalochordate Branchiostoma lanceolatum (see Figure | + | mNeonGreen is a yellow-green fluorescent protein and is derived from a tetrameric fluorescent protein from cephalochordate Branchiostoma lanceolatum (see Figure 2). mNeonGreen is the brightest monomeric green or yellow fluorescent protein yet described and is an excellent fluorescence resonance energy transfer (FRET) acceptor for the newest cyan fluorescent proteins [1]. |
[[File:TU_Eindhoven_mNeonGreen_3D-structure.png]] | [[File:TU_Eindhoven_mNeonGreen_3D-structure.png]] | ||
| − | ''Figure | + | ''Figure 2: Schamatic 3D structure of mNeonGreen'' |
| + | |||
| + | == Sequence comparison of mNeonGreen with EGFP == | ||
| + | |||
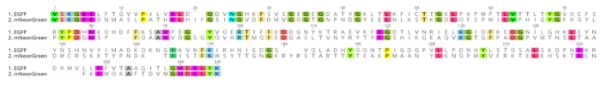
| + | mNeonGreen consists of 237 amino acids, which translates into 26.6 kDa molecular weight. mNeonGreen is evolutionarily distant from jellyfish-derived fluorescent proteins. At sequence level, mNeonGreen shares just 20-25% sequence identity with common GFP derivatives. | ||
| + | |||
| + | [[File:Athens2022_sequence_comparison_mNeonGreen.png]] | ||
| + | |||
| + | ''Figure 3: Sequence comparison of mNeonGreen with EGFP '' | ||
== Gene Design == | == Gene Design == | ||
mNeonGreen was characterized by Shaner et al [1]. We inserted mNeonGreen in the pETDuet-1 vector together with OmpX and a BamHI-linker (see Figure 2) [https://parts.igem.org/Part:BBa_K1761001]. This linker is a 213 bp long flexible GGSGGS linker and by using the restriction enzyme BamHI, the linker can become 45 bp shorter. | mNeonGreen was characterized by Shaner et al [1]. We inserted mNeonGreen in the pETDuet-1 vector together with OmpX and a BamHI-linker (see Figure 2) [https://parts.igem.org/Part:BBa_K1761001]. This linker is a 213 bp long flexible GGSGGS linker and by using the restriction enzyme BamHI, the linker can become 45 bp shorter. | ||
| − | The BamHI-linker is inspired by the article "Quantitative Understanding of the Energy Transfer between Fluorescent Proteins Connected via Flexible Peptide Liners" by T.H. Evers et al from 29 August 2006 | + | The BamHI-linker is inspired by the article "Quantitative Understanding of the Energy Transfer between Fluorescent Proteins Connected via Flexible Peptide Liners" by T.H. Evers et al from 29 August 2006 [2]. |
[[File:TU_Eindhoven_Construct_OmpX_mNeonGreen.png]] | [[File:TU_Eindhoven_Construct_OmpX_mNeonGreen.png]] | ||
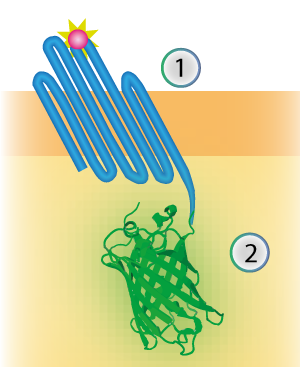
| − | ''Figure | + | ''Figure 4: Schematical overview of the expressed OmpX (1) with mNeonGreen (2) in the outer membrane of E.coli.'' |
| − | + | ||
== Sequence == | == Sequence == | ||
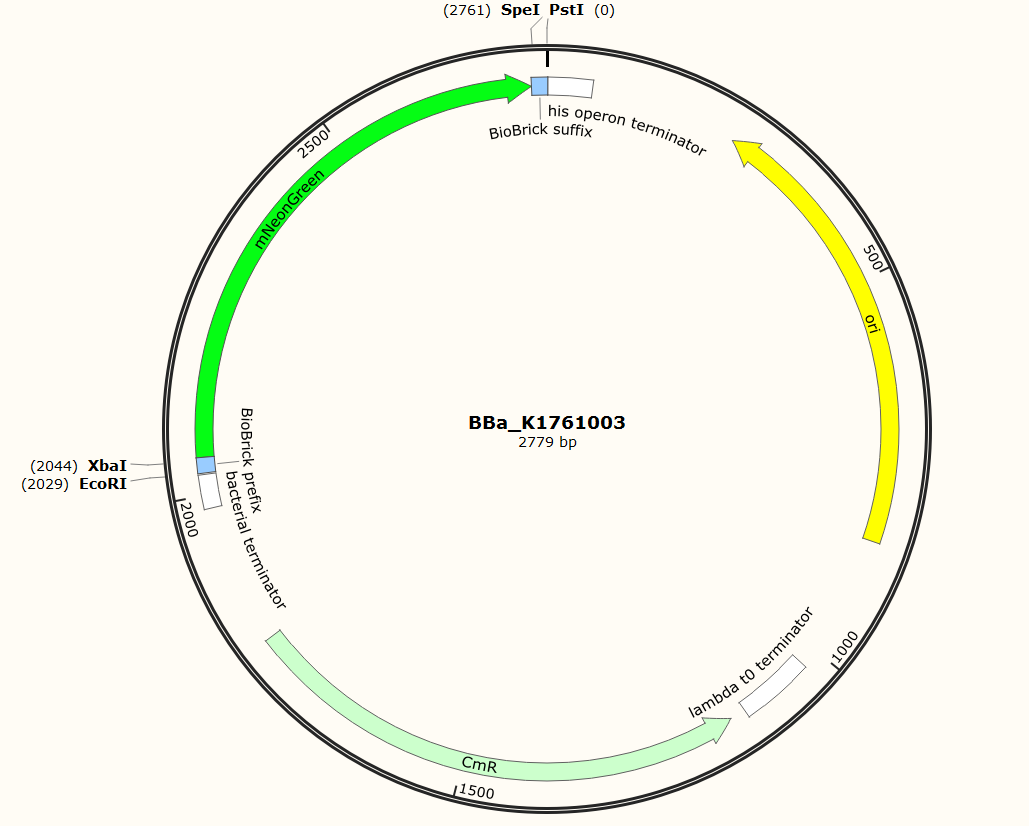
The sequence of our mTurquoise2 part (see Figure 3) has been verified by sequencing at StarSeq. It contains the prefix and suffix with the correct restriction sites (EcoRI, XbaI, SpeI and PstI). mNeonGreen is 711 bp long. | The sequence of our mTurquoise2 part (see Figure 3) has been verified by sequencing at StarSeq. It contains the prefix and suffix with the correct restriction sites (EcoRI, XbaI, SpeI and PstI). mNeonGreen is 711 bp long. | ||
| − | [[File:TU_Eindhoven_mNeonGreen_SnapGene.png]] | + | [[File:TU_Eindhoven_mNeonGreen_SnapGene.png|800px]] |
| − | ''Figure | + | ''Figure 5: Snapgene plasmid overview of the BioBrick part BBa_K1761003. It shows the pSB1C3 vector with the prefix (containing the restriction sites EcoRI and XbaI), mNeonGreen and the suffix (containting the restiction sites SpeI and PstI).'' |
<!-- --> | <!-- --> | ||
| Line 34: | Line 44: | ||
== Characterization == | == Characterization == | ||
| − | + | mNeonGreen was characterized by fusing it to an outer membrane protein (see below) as well as by expressing it on itself in the cytosol. For the expression of mNeonGreen in the cytosol, mNeonGreen was inserted after the constitutive promotor J23101. Even though no RBS was introduced into the composite part, the cells still showed fluorescence which could be measured with the Cary Eclipse spectrofluorometer for both an in vivo excitation scan and a in vivo emission scan. | |
| − | For | + | |
| + | |||
| + | == In vivo excitation scan == | ||
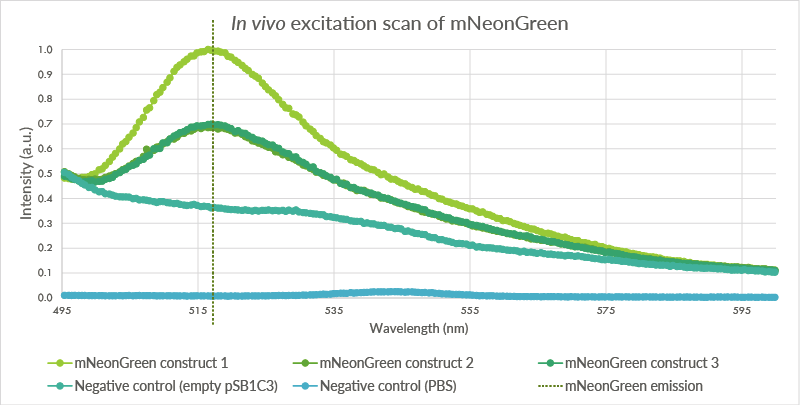
| + | An in vivo excitation scan was conducted for the mNeonGreen containing cells (see Figure 4). Biological triplicates were measured. The cells were measured at an OD of 2.0 with the Cary Eclipse spectrofluorometer and showed a peak at approximately 517 nm. The first negative control, BL21(DE3) cells transformed with an empty PSB1C3 vector, showed no such fluorescence even though light intensity was measured. The second negative control showed almost no light intensity, indicating that the light intensity for the first negative control is a result of light scattering. | ||
| + | |||
| + | [[File:TU_Eindhoven_mNeonGreen_ExcitationScan.png]] | ||
| + | |||
| + | ''Figure 6: The pSB1C3 vector containing mNeonGreen vector was transformed into BL21(DE3) cells. These cells were measured at an OD of 2.0. Three biological triplicates were obtained which all showed peaks at approximately 517 nm. The first negative control, cells with an empty pSB1C3 plasmid, do not show such a peak. The second negative control, pure PBS, showed a very low intensity.'' | ||
| + | |||
| + | |||
| + | == In vivo emission scan == | ||
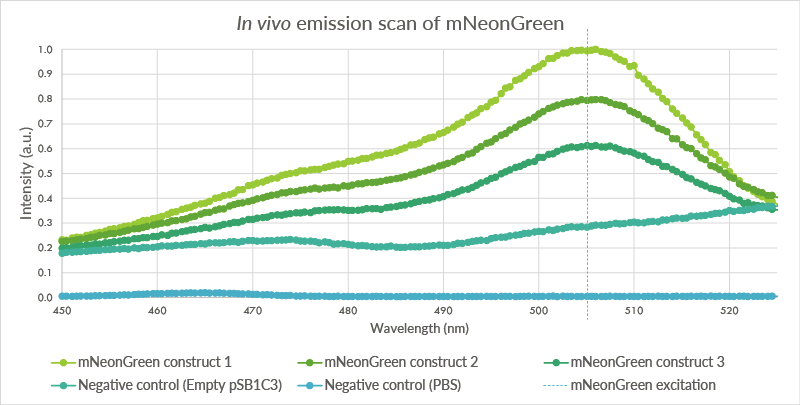
| + | An in vivo emission scan was conducted for the mNeonGreen containg cells (see Figure 5). The test was conducted for the same biological triplicate as the in vivo excitation scan. Cells were measured at an OD of 2.0 with the Cary Eclipse spectrofluorometer and showed an excitation peak at approximately 506 nm. | ||
| + | |||
| + | [[File:TU_Eindhoven_mNeonGreen_EmissionScan.png]] | ||
| + | |||
| + | ''Figure 7: An in vivo emissions scan was conducted for mNeonGreen in the BL21(DE3) cells. The cells containing mNeonGreen show a peak at 506 nm. The first negative control, BL21(DE3) cells with the empty pSB1C3 plasmid do not show such a peak. The second negative control, pure PBS, shows almost no fluorescence. The higher level of fluorescence of the BL21(DE3) cells is possibly caused by autofluorescence.'' | ||
| + | |||
| + | |||
| + | == Spectra of the mNeonGreen biological triplicate == | ||
| + | For the mNeonGreen-containing BL21(DE3) cells, emission and excitation spectra were obtained. These spectra are shown below in Figure 8, 9 and 10. | ||
| + | |||
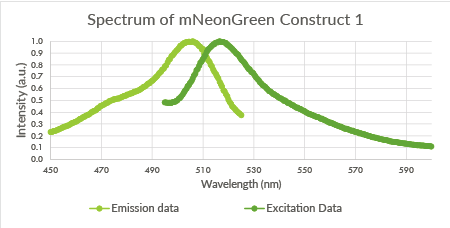
| + | [[File:TU_Eindhoven_mNeonGreen_Construct1.png]] | ||
| + | |||
| + | ''Figure 8: Spectrum of mNeonGreen and constitutive promotor J23101, Construct 1.'' | ||
| + | |||
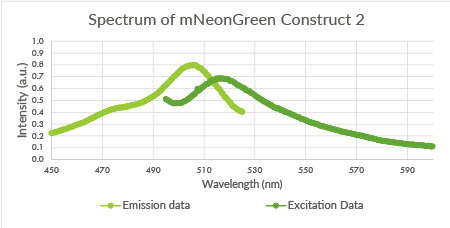
| + | [[File:TU_Eindhoven_mNeonGreen_Construct2.png]] | ||
| + | |||
| + | ''Figure 9: Spectrum of mNeonGreen and constitutive promotor J23101, Construct 2.'' | ||
| + | |||
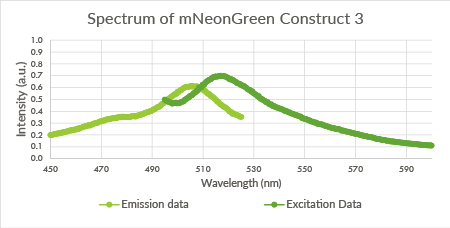
| + | [[File:TU_Eindhoven_mNeonGreen_Construct3.png]] | ||
| + | |||
| + | ''Figure 10: Spectrum of mNeonGreen and constitutive promotor J23101, Construct 3.'' | ||
| + | |||
| + | |||
| + | == Measurement with the Tecan Safire2 platereader == | ||
| + | In addition to measurement with the spectrofluorometer, we measured fluorescence of the mNeonGreen-containing cells with the Tecan Safire2 plate-reader. The cells were diluted to an OD of approximately 1.5 before they were loaded in the wells. For the three biological replicates, three technical replicates were measured. The average of these three technical replicates was used for the graph in Figure 9. | ||
| + | |||
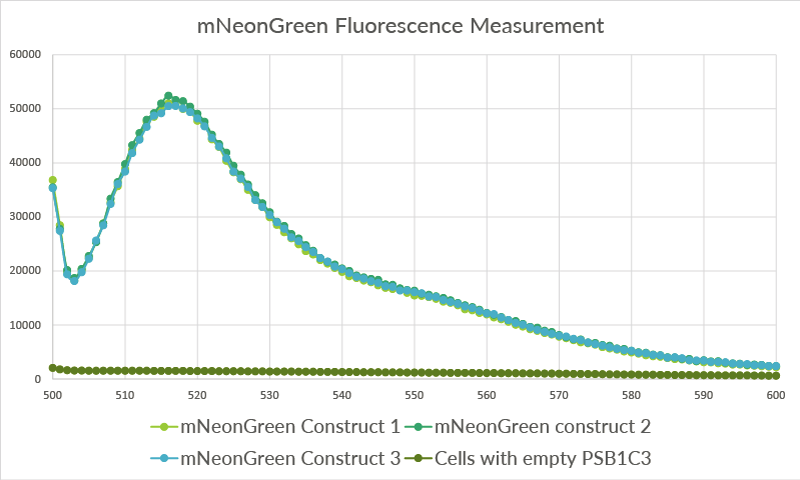
| + | [[File:TU_Eindhoven_mNeonGreen_Safire2.png]] | ||
| + | |||
| + | ''Figure 11: The data for the three biological replicates shows high intensity peaks at approximately 517 nm. Three technical replicates for each of the biological replicates were measured. The average of these three technical replicates was used for the shown spectra. Excitation was performed at 480 nm with a bandwidth of 5nm. Emission was measured from 500 nm till 600 nm with a bandwidth size of 7 nm.'' | ||
| + | |||
| + | == Characterization part of a construct == | ||
| + | As already mentioned, mNeonGreen was also characterized by fusing it to an outer membrane protein by preforming a fluorescence assay. | ||
| + | For all the experiments below, we used the following vectors: pETDuet-1 with a construct inserted (OmpX + intracellular protein) and pEVOL-pAzF (tRNA + tRNA synthetase). Both vectors were transformed into BL21(DE3). The expression of the construct was introduced by adding arabinose, IPTG and the unnatural amino acid. | ||
== Fluorescence Confirmation == | == Fluorescence Confirmation == | ||
| − | To confirm whether mNeonGreen is present in the bacteria, a fluorescence assay was performed. Excitation took place at a wavelength of 480 nm with a laser. Emission was read out at 517 nm. From this experiment, it can be concluded that mNeonGreen is present and works (see Figure | + | To confirm whether mNeonGreen is present in the bacteria, a fluorescence assay was performed. Excitation took place at a wavelength of 480 nm with a laser. Emission was read out at 517 nm. From this experiment, it can be concluded that mNeonGreen is present and works (see Figure 12). This was also tested with both OmpX – mNeonGreen and OmpX – NanoLuc inserted, which gave the same results (see Figure 13). |
For more information about how to perform a fluorescence assay, see our Protocol Page [http://2015.igem.org/Team:TU_Eindhoven/Project/Protocols]. | For more information about how to perform a fluorescence assay, see our Protocol Page [http://2015.igem.org/Team:TU_Eindhoven/Project/Protocols]. | ||
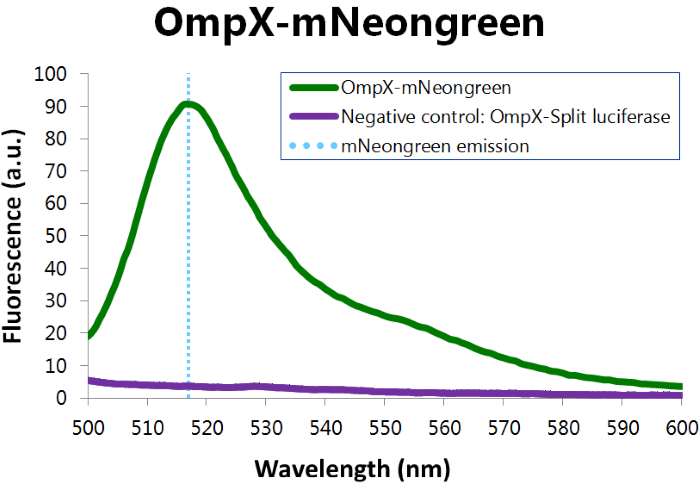
[[File:TU_Eindhoven_Results_Fluorescence_mNeonGreen.png]] | [[File:TU_Eindhoven_Results_Fluorescence_mNeonGreen.png]] | ||
| − | ''Figure | + | ''Figure 12: Fluorescence results of OmpX - mNeonGreen.'' |
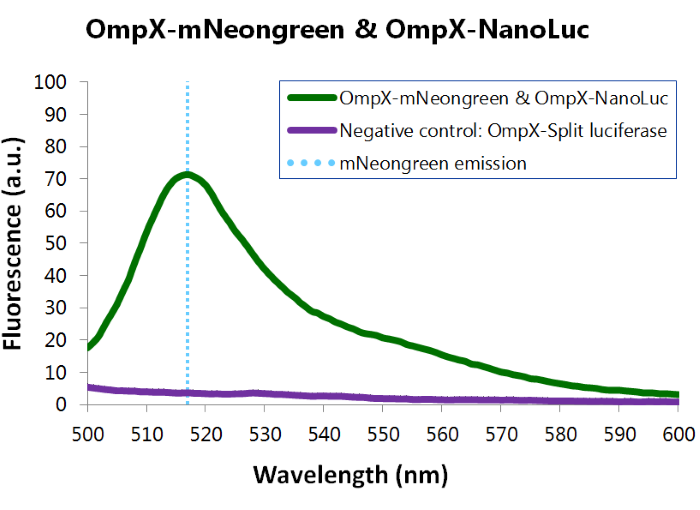
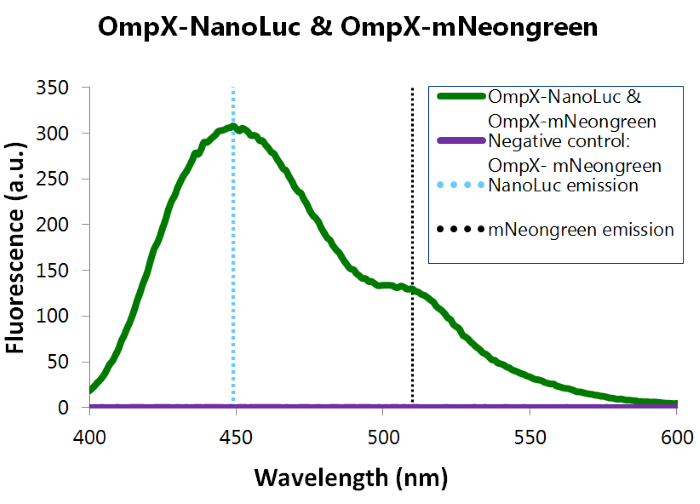
[[File:TU_Eindhoven_Results_Fluorescence_mNeonGreen_and_NanoLuc.png]] | [[File:TU_Eindhoven_Results_Fluorescence_mNeonGreen_and_NanoLuc.png]] | ||
| − | ''Figure | + | ''Figure 13: Fluorescence results of OmpX -mNeonGreen and OmpX - NanoLuc.'' |
| + | == Bioluminescence Confirmation == | ||
| + | To confirm whether mNeonGreen forms a good BRET (Bioluminescence resonance energy transfer) couple with NanoLuc, a bioluminescence measurement was performed (see Figure 12). In this experiment OmpX-NanoLuc's & OmpX-mNeonGreen's presence in the cells were tested by adding Nano-Glo substrate to the cells and measuring bioluminescence with the spectrophotometer. Figure 8 shows that OmpX-NanoLuc shows a peak characteristic for NanoLuc, indicating NanoLuc's presence within the cells. Moreover, the spectrogram shows a distinct shoulder near 517 nm, the emission wavelength of mNeongreen. Since no laser was used, excitation of mNeongreen can only be accomplished by NanoLuc so BRET (Bioluminescence resonance energy transfer) occured. This signal is measured when no click reaction is performed on the complex, meaning that this can be seen as the background noise of the sensor of iGEM team TU Eindhoven 2015. For more information about how to perform a bioluminescence measurement, see our Protocol Page [http://2015.igem.org/Team:TU_Eindhoven/Project/Protocols]. | ||
| + | |||
| + | [[File:TU_Eindhoven_results_bioluminiscence_neongreen_and_NanoLuc.png]] | ||
| + | |||
| + | ''Figure 14: Bioluminiscence results NanoLuc and mNeonGreen.'' | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
| Line 56: | Line 114: | ||
<!-- --> | <!-- --> | ||
| + | ==Improvement - Secreting Chemokine with mNeonGreen Tag== | ||
| + | MIT iGEM 2019 Team | ||
| + | We improved mNeonGreen (BBa_K1761003) by using it as a fusion protein tag and attaching it to the IL-8 gene in the form of BBa_K2957002 (IL8 with NeonGreen tag). We believe we improved this part for the following reasons: | ||
| + | <ul><li>We improved this part by proving its use as a <b>fusion fluorescent protein tag</b> to the IL-8 chemokine and CCL5 chemokine. This allows it to visually be tracked in the cell. </li> | ||
| + | <li>We proved that it can be secreted out of the cell carried by a <b>chemokine with a natural secretion tag </b>. (see secretion results on composite parts page, BBa_K2957007) </li> | ||
| + | <i>see https://2019.igem.org/Team:MIT/Composite_Part </i> | ||
| + | </ul> | ||
== References == | == References == | ||
| Line 61: | Line 126: | ||
[2] T.H. Evers et al, “Quantitative Understanding of the Energy Transfer between Fluorescent Proteins Connected via Flexible Peptide Linkers.,” Biochemistry, vol. 45, no. 44, pp. 13183-92, Nov. 2006. | [2] T.H. Evers et al, “Quantitative Understanding of the Energy Transfer between Fluorescent Proteins Connected via Flexible Peptide Linkers.,” Biochemistry, vol. 45, no. 44, pp. 13183-92, Nov. 2006. | ||
| + | |||
| + | [3] mNeonGreen, FPbase :: The Fluorescent Protein Database, https://www.fpbase.org/protein/mneongreen/ October 12 2022 | ||
| + | |||
| + | [4] Bright Monomeric Fluorescent Proteins: mNeonGreen, mTFP1, and mWasabi, Jennifer Tsang, Addgene Blog October 12 2022 | ||
| + | |||
| + | [5] Hostettler L, Grundy L, Käser-Pébernard S, Wicky C, Schafer WR, Glauser DA. The Bright Fluorescent Protein mNeonGreen Facilitates Protein Expression Analysis In Vivo. G3 (Bethesda). 2017 Feb 9;7(2):607-615. doi: 10.1534/g3.116.038133. PMID: 28108553; PMCID: PMC5295605. | ||
| + | |||
| + | [6] Shaner, N., Lambert, G., Chammas, A. et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods 10, 407–409 (2013). https://doi.org/10.1038/nmeth.2413 | ||
| + | |||
| + | [7] mNeonGreen vs GFP, https://www.ptglab.com/news/blog/mneongreen-vs-gfp/?fbclid=IwAR25JJISJurn_cDT4b0aRc3z2fpFbfk1OEvFHOgRLqnHOtCcbQdh5MdD0sE, October 12 2022 | ||
Latest revision as of 20:01, 13 October 2022
mNeonGreen
mNeonGreen is a yellow-green fluorescent protein. For a fluorescene measurement, the mNeonGreen fluorophore can be excitated with a laser with a wavelenght of 480 nm and readed out at 517 nm.
Figure 1: Excitation and Emission wavelengths of mNeonGreen
Usage and Biology
mNeonGreen is a yellow-green fluorescent protein and is derived from a tetrameric fluorescent protein from cephalochordate Branchiostoma lanceolatum (see Figure 2). mNeonGreen is the brightest monomeric green or yellow fluorescent protein yet described and is an excellent fluorescence resonance energy transfer (FRET) acceptor for the newest cyan fluorescent proteins [1].
Figure 2: Schamatic 3D structure of mNeonGreen
Sequence comparison of mNeonGreen with EGFP
mNeonGreen consists of 237 amino acids, which translates into 26.6 kDa molecular weight. mNeonGreen is evolutionarily distant from jellyfish-derived fluorescent proteins. At sequence level, mNeonGreen shares just 20-25% sequence identity with common GFP derivatives.
Figure 3: Sequence comparison of mNeonGreen with EGFP
Gene Design
mNeonGreen was characterized by Shaner et al [1]. We inserted mNeonGreen in the pETDuet-1 vector together with OmpX and a BamHI-linker (see Figure 2) [1]. This linker is a 213 bp long flexible GGSGGS linker and by using the restriction enzyme BamHI, the linker can become 45 bp shorter. The BamHI-linker is inspired by the article "Quantitative Understanding of the Energy Transfer between Fluorescent Proteins Connected via Flexible Peptide Liners" by T.H. Evers et al from 29 August 2006 [2].
Figure 4: Schematical overview of the expressed OmpX (1) with mNeonGreen (2) in the outer membrane of E.coli.
Sequence
The sequence of our mTurquoise2 part (see Figure 3) has been verified by sequencing at StarSeq. It contains the prefix and suffix with the correct restriction sites (EcoRI, XbaI, SpeI and PstI). mNeonGreen is 711 bp long.
Figure 5: Snapgene plasmid overview of the BioBrick part BBa_K1761003. It shows the pSB1C3 vector with the prefix (containing the restriction sites EcoRI and XbaI), mNeonGreen and the suffix (containting the restiction sites SpeI and PstI).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
mNeonGreen was characterized by fusing it to an outer membrane protein (see below) as well as by expressing it on itself in the cytosol. For the expression of mNeonGreen in the cytosol, mNeonGreen was inserted after the constitutive promotor J23101. Even though no RBS was introduced into the composite part, the cells still showed fluorescence which could be measured with the Cary Eclipse spectrofluorometer for both an in vivo excitation scan and a in vivo emission scan.
In vivo excitation scan
An in vivo excitation scan was conducted for the mNeonGreen containing cells (see Figure 4). Biological triplicates were measured. The cells were measured at an OD of 2.0 with the Cary Eclipse spectrofluorometer and showed a peak at approximately 517 nm. The first negative control, BL21(DE3) cells transformed with an empty PSB1C3 vector, showed no such fluorescence even though light intensity was measured. The second negative control showed almost no light intensity, indicating that the light intensity for the first negative control is a result of light scattering.
Figure 6: The pSB1C3 vector containing mNeonGreen vector was transformed into BL21(DE3) cells. These cells were measured at an OD of 2.0. Three biological triplicates were obtained which all showed peaks at approximately 517 nm. The first negative control, cells with an empty pSB1C3 plasmid, do not show such a peak. The second negative control, pure PBS, showed a very low intensity.
In vivo emission scan
An in vivo emission scan was conducted for the mNeonGreen containg cells (see Figure 5). The test was conducted for the same biological triplicate as the in vivo excitation scan. Cells were measured at an OD of 2.0 with the Cary Eclipse spectrofluorometer and showed an excitation peak at approximately 506 nm.
Figure 7: An in vivo emissions scan was conducted for mNeonGreen in the BL21(DE3) cells. The cells containing mNeonGreen show a peak at 506 nm. The first negative control, BL21(DE3) cells with the empty pSB1C3 plasmid do not show such a peak. The second negative control, pure PBS, shows almost no fluorescence. The higher level of fluorescence of the BL21(DE3) cells is possibly caused by autofluorescence.
Spectra of the mNeonGreen biological triplicate
For the mNeonGreen-containing BL21(DE3) cells, emission and excitation spectra were obtained. These spectra are shown below in Figure 8, 9 and 10.
Figure 8: Spectrum of mNeonGreen and constitutive promotor J23101, Construct 1.
Figure 9: Spectrum of mNeonGreen and constitutive promotor J23101, Construct 2.
Figure 10: Spectrum of mNeonGreen and constitutive promotor J23101, Construct 3.
Measurement with the Tecan Safire2 platereader
In addition to measurement with the spectrofluorometer, we measured fluorescence of the mNeonGreen-containing cells with the Tecan Safire2 plate-reader. The cells were diluted to an OD of approximately 1.5 before they were loaded in the wells. For the three biological replicates, three technical replicates were measured. The average of these three technical replicates was used for the graph in Figure 9.
Figure 11: The data for the three biological replicates shows high intensity peaks at approximately 517 nm. Three technical replicates for each of the biological replicates were measured. The average of these three technical replicates was used for the shown spectra. Excitation was performed at 480 nm with a bandwidth of 5nm. Emission was measured from 500 nm till 600 nm with a bandwidth size of 7 nm.
Characterization part of a construct
As already mentioned, mNeonGreen was also characterized by fusing it to an outer membrane protein by preforming a fluorescence assay. For all the experiments below, we used the following vectors: pETDuet-1 with a construct inserted (OmpX + intracellular protein) and pEVOL-pAzF (tRNA + tRNA synthetase). Both vectors were transformed into BL21(DE3). The expression of the construct was introduced by adding arabinose, IPTG and the unnatural amino acid.
Fluorescence Confirmation
To confirm whether mNeonGreen is present in the bacteria, a fluorescence assay was performed. Excitation took place at a wavelength of 480 nm with a laser. Emission was read out at 517 nm. From this experiment, it can be concluded that mNeonGreen is present and works (see Figure 12). This was also tested with both OmpX – mNeonGreen and OmpX – NanoLuc inserted, which gave the same results (see Figure 13). For more information about how to perform a fluorescence assay, see our Protocol Page [http://2015.igem.org/Team:TU_Eindhoven/Project/Protocols].
Figure 12: Fluorescence results of OmpX - mNeonGreen.
Figure 13: Fluorescence results of OmpX -mNeonGreen and OmpX - NanoLuc.
Bioluminescence Confirmation
To confirm whether mNeonGreen forms a good BRET (Bioluminescence resonance energy transfer) couple with NanoLuc, a bioluminescence measurement was performed (see Figure 12). In this experiment OmpX-NanoLuc's & OmpX-mNeonGreen's presence in the cells were tested by adding Nano-Glo substrate to the cells and measuring bioluminescence with the spectrophotometer. Figure 8 shows that OmpX-NanoLuc shows a peak characteristic for NanoLuc, indicating NanoLuc's presence within the cells. Moreover, the spectrogram shows a distinct shoulder near 517 nm, the emission wavelength of mNeongreen. Since no laser was used, excitation of mNeongreen can only be accomplished by NanoLuc so BRET (Bioluminescence resonance energy transfer) occured. This signal is measured when no click reaction is performed on the complex, meaning that this can be seen as the background noise of the sensor of iGEM team TU Eindhoven 2015. For more information about how to perform a bioluminescence measurement, see our Protocol Page [http://2015.igem.org/Team:TU_Eindhoven/Project/Protocols].
Figure 14: Bioluminiscence results NanoLuc and mNeonGreen.
Improvement - Secreting Chemokine with mNeonGreen Tag
MIT iGEM 2019 Team We improved mNeonGreen (BBa_K1761003) by using it as a fusion protein tag and attaching it to the IL-8 gene in the form of BBa_K2957002 (IL8 with NeonGreen tag). We believe we improved this part for the following reasons:
- We improved this part by proving its use as a fusion fluorescent protein tag to the IL-8 chemokine and CCL5 chemokine. This allows it to visually be tracked in the cell.
- We proved that it can be secreted out of the cell carried by a chemokine with a natural secretion tag . (see secretion results on composite parts page, BBa_K2957007)
References
[1] N.C. Shaner et al, “A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum.,” Nature Methods, vol. 10, no. 5, pp. 407-409, Mar. 2013.
[2] T.H. Evers et al, “Quantitative Understanding of the Energy Transfer between Fluorescent Proteins Connected via Flexible Peptide Linkers.,” Biochemistry, vol. 45, no. 44, pp. 13183-92, Nov. 2006.
[3] mNeonGreen, FPbase :: The Fluorescent Protein Database, https://www.fpbase.org/protein/mneongreen/ October 12 2022
[4] Bright Monomeric Fluorescent Proteins: mNeonGreen, mTFP1, and mWasabi, Jennifer Tsang, Addgene Blog October 12 2022
[5] Hostettler L, Grundy L, Käser-Pébernard S, Wicky C, Schafer WR, Glauser DA. The Bright Fluorescent Protein mNeonGreen Facilitates Protein Expression Analysis In Vivo. G3 (Bethesda). 2017 Feb 9;7(2):607-615. doi: 10.1534/g3.116.038133. PMID: 28108553; PMCID: PMC5295605.
[6] Shaner, N., Lambert, G., Chammas, A. et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods 10, 407–409 (2013). https://doi.org/10.1038/nmeth.2413
[7] mNeonGreen vs GFP, https://www.ptglab.com/news/blog/mneongreen-vs-gfp/?fbclid=IwAR25JJISJurn_cDT4b0aRc3z2fpFbfk1OEvFHOgRLqnHOtCcbQdh5MdD0sE, October 12 2022