Difference between revisions of "Part:BBa K1351017"
| (10 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1351017 short</partinfo> | <partinfo>BBa_K1351017 short</partinfo> | ||
| − | This gene codes for the immunity against | + | This gene codes for the immunity against the cannibalism toxin SDP of ''Bacillus subtilis''. The ''sdpI'' gene is regulated by a separate promoter (PsdpRI) which is responsible for the operon sdpRI. Cannibalsim toxins are produced by ''B. subtilis'' mainly in mid to late stationary phase when nutrients are limited. |
| + | We wanted to evaluate the immunity in order to later on interfere with it for our [http://2014.igem.org/Team:LMU-Munich/Project/Bakillus suicide module]. | ||
| + | For more details about our interference strategies please check our antisense RNA [https://parts.igem.org/Part:BBa_K1351019 BBa_K1351019] and our small RNA fsrA [https://parts.igem.org/Part:BBa_K1351022 BBa_K1351022]. | ||
| + | <br><br> | ||
Prefix: TGCAGAATTCGCGGCCGCTTCTAGAGTAAGGAGGAACTACT<br> | Prefix: TGCAGAATTCGCGGCCGCTTCTAGAGTAAGGAGGAACTACT<br> | ||
Suffix: TACTAGTAGCGGCCGCTGCAGGCT | Suffix: TACTAGTAGCGGCCGCTGCAGGCT | ||
<br> | <br> | ||
Results: [http://2014.igem.org/Team:LMU-Munich/Results LMU-2014-Results] | Results: [http://2014.igem.org/Team:LMU-Munich/Results LMU-2014-Results] | ||
| − | + | <br> | |
This part is used in the 2014 LMU-Munich iGEM project [http://2014.igem.org/Team:LMU-Munich BaKillus]. | This part is used in the 2014 LMU-Munich iGEM project [http://2014.igem.org/Team:LMU-Munich BaKillus]. | ||
| − | + | <br><br> | |
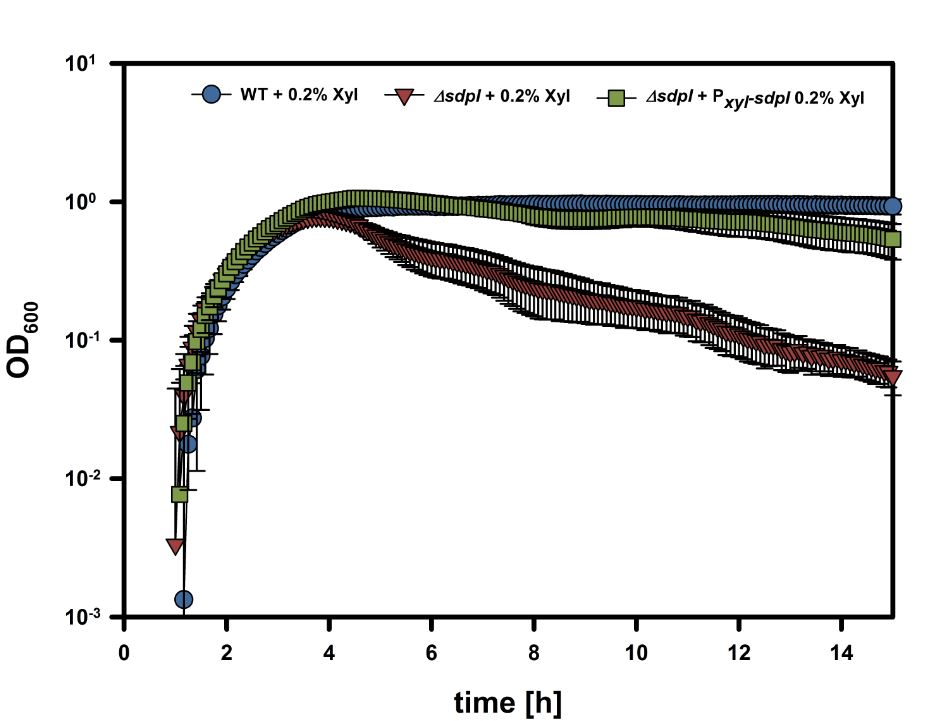
First results of this BioBrick are depicted in the following graph: | First results of this BioBrick are depicted in the following graph: | ||
[[File:LMU14Sdpi.PNG|600px]] | [[File:LMU14Sdpi.PNG|600px]] | ||
| + | |||
| + | The graph shows growth curves of ''Bacillus subtilis'' W168 wildtype, a ''sdpI'' mutant strain as well as a ''sdpI'' mutant strain carrying our BioBrick [https://parts.igem.org/wiki/index.php?title=Part:BBa_K1351017 BBa_K1351017]. | ||
| + | The results were performed with our protocoll: [https://static.igem.org/mediawiki/2014/c/c7/LMU-Munich14_Luminescence_Assay.pdf Lumi Assay] in MCSE and consist of three replicas of three measurements. The induction was performed with 0.2% xylose. | ||
| + | These results clearly show that our BioBrick [https://parts.igem.org/wiki/index.php?title=Part:BBa_K1351017 BBa_K1351017] is functional and recovers wild type growth behavior. | ||
| + | <br><br> | ||
Latest revision as of 15:58, 30 October 2014
SdpI with RBS: Immunity against the cannibalsim toxin sdpC of B. subtilis
This gene codes for the immunity against the cannibalism toxin SDP of Bacillus subtilis. The sdpI gene is regulated by a separate promoter (PsdpRI) which is responsible for the operon sdpRI. Cannibalsim toxins are produced by B. subtilis mainly in mid to late stationary phase when nutrients are limited. We wanted to evaluate the immunity in order to later on interfere with it for our [http://2014.igem.org/Team:LMU-Munich/Project/Bakillus suicide module]. For more details about our interference strategies please check our antisense RNA BBa_K1351019 and our small RNA fsrA BBa_K1351022.
Prefix: TGCAGAATTCGCGGCCGCTTCTAGAGTAAGGAGGAACTACT
Suffix: TACTAGTAGCGGCCGCTGCAGGCT
Results: [http://2014.igem.org/Team:LMU-Munich/Results LMU-2014-Results]
This part is used in the 2014 LMU-Munich iGEM project [http://2014.igem.org/Team:LMU-Munich BaKillus].
First results of this BioBrick are depicted in the following graph:
The graph shows growth curves of Bacillus subtilis W168 wildtype, a sdpI mutant strain as well as a sdpI mutant strain carrying our BioBrick BBa_K1351017.
The results were performed with our protocoll: Lumi Assay in MCSE and consist of three replicas of three measurements. The induction was performed with 0.2% xylose.
These results clearly show that our BioBrick BBa_K1351017 is functional and recovers wild type growth behavior.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]