Difference between revisions of "Part:BBa K1351036"
| (One intermediate revision by the same user not shown) | |||
| Line 19: | Line 19: | ||
[[File:LMU14 Cap2p3.png|600px]] | [[File:LMU14 Cap2p3.png|600px]] | ||
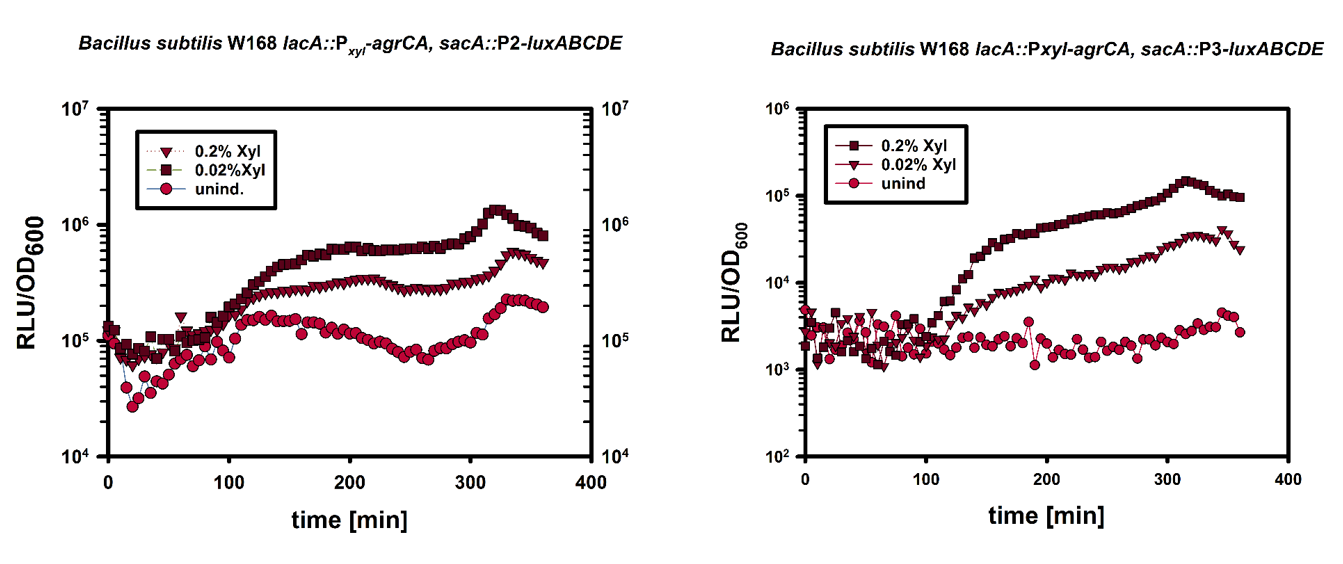
| − | Luminescence of ''B. subtilis'' P<sub>''xylA''</sub> - ''agrCA'' / P2 | + | Luminescence of ''B. subtilis'' P<sub>''xylA''</sub> - ''agrCA'' / [https://parts.igem.org/Part:BBa_K1351037 P2]-''lux'' and [https://parts.igem.org/Part:BBa_K1351038 P3]-''lux'' was measured in MCSE induced with 0, 0.02, 0.2 % xylose. This plot shows the luminescence readout normalised over OD600 over the course of 6hours. The higher output in xylose induced samples shows the successfull expression of ''agrCA'' in ''B. subtilis''. |
| − | |||
| − | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
Latest revision as of 09:21, 27 October 2014
Quorum sensing two component system (AIP-II sensing histidine kinase agrC, response regulator agrA)
Quorum sensing two component system derived from Staphylococcus aureus N315. This part consists of the AIP-II sensing histidine kinase AgrC and the response regulator AgrA. Upon detection of autoinducing peptide II (AIP-II), AgrC phosphorylates AgrA, which then bindes to distinct binding sites and activates the P2 and P3 promoters. This part should only work for AIP-II. Other AIPs should lead to inhibition.
This part was generated in a modified version of RFC25, where a strong Shine Dalgarno Sequence (SD) is included, and has the following prefix and suffix:
| prefix with EcoRI, NotI, XbaI, SD and NgoMIV: | GAATTCGCGGCCGCTTCTAGAGTAAGGAGGAGCCGGC |
| suffix with AgeI, SpeI, NotI and PstI: | ACCGGTTAATACTAGTAGCGGCCGCTGCAG |
Sites of restriction enzymes generating compatible overhangs have the same color:
EcoRI and PstI in blue, NotI in green, XbaI and SpeI in red, NgoMIV and AgeI in orange. Shine-Dalgarno sequence and stop codons are underlined.
This part is used in the 2014 LMU-Munich iGEM project [http://2014.igem.org/Team:LMU-Munich BaKillus].
Luminescence of B. subtilis PxylA - agrCA / P2-lux and P3-lux was measured in MCSE induced with 0, 0.02, 0.2 % xylose. This plot shows the luminescence readout normalised over OD600 over the course of 6hours. The higher output in xylose induced samples shows the successfull expression of agrCA in B. subtilis.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1139
Illegal AgeI site found at 1114 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 475