Difference between revisions of "Part:BBa K1378031"
(→Result) |
|||
| (6 intermediate revisions by the same user not shown) | |||
| Line 53: | Line 53: | ||
<partinfo>BBa_K1378031 parameters</partinfo> | <partinfo>BBa_K1378031 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | |||
| + | |||
| + | ==Team NYMU-Taipei 2017: Successfully construct suicide mechanism by T4 phage== | ||
| + | |||
| + | ===Improvement=== | ||
| + | The gene sequences and the structures of holin in different species of bacteriophages are also different. In NYMU-Taipei 2017 team’s project, we choose the holin from enterobacteria phage T4. The T4 holin we used is an iGEM released part, BBa_K112000. We combine T4 holin with a lactose-induced promoter (BBa_R0010), a ribosome binding site (BBa_B0034) and a double terminator (BBa_B0010 and BBa_B0012). Therefore, holin can be induced by lactose and regulate the suicide mechanism. Moreover, we construct T4 holin and T4 endolysin together as a functional suicide mechanism. Besides, in our team NYMU-Taipei 2017 project, we also put the suicide mechanism into practice by constructing them with NrtA. | ||
| + | |||
| + | |||
| + | ===Result=== | ||
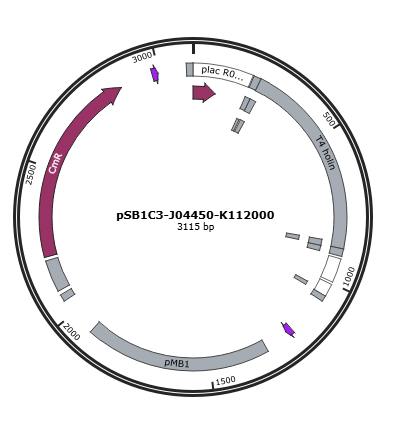
| + | Figure 1 is the gene map of T4 holin construct, pSB1C3-J04450-K112000. <br> | ||
| + | |||
| + | <b>Figure 1 </b> | ||
| + | |||
| + | [[File:T--NYMU-Taipei--Holin.png|400px]] | ||
| + | |||
| + | |||
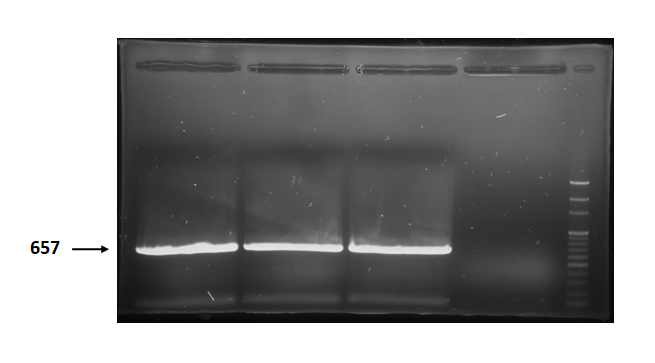
| + | Figure 2 is electrophoresis result of holin (from BBa_K112000) PCR product. | ||
| + | The marker is 100bp. The length is 657bp as expected. <br> | ||
| + | |||
| + | <b>Figure 2 </b> | ||
| + | |||
| + | [[File:T--NYMU-Taipei--Holin PCR 657bp.png|400px]] | ||
| + | |||
| + | |||
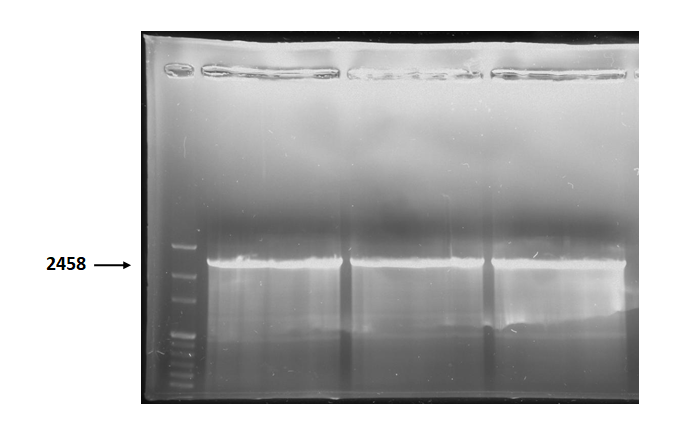
| + | Figure 3 is electrophoresis result of holin backbone (from BBa_J04450) PCR product. The marker is 1kb. The length is 2458bp as expected. <br> | ||
| + | |||
| + | <b>Figure 3 </b> | ||
| + | |||
| + | [[File:T--NYMU-Taipei--Holin BB PCR 2458bp.png|400px]] | ||
| + | |||
| + | |||
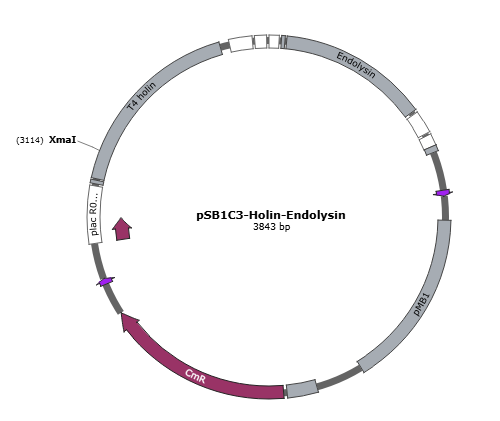
| + | Figure 4 is the gene map of suicide mechanism construct, pSB1C3- T4 holin- T4 endolysin. <br> | ||
| + | |||
| + | <b>Figure 4 </b> | ||
| + | |||
| + | [[File:T--NYMU-Taipei--Holin-Endolysin XmaI.png|400px]] | ||
| + | |||
| + | |||
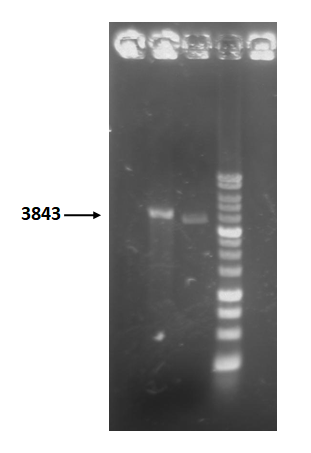
| + | Figure 5 is restriction enzyme check electrophoresis result of T4 holin- T4 endolysin construct. | ||
| + | The marker is 1kb. We use XmaI to digest the sample. The total length is 3843bp as expected. <br> | ||
| + | |||
| + | <b>Figure 5 </b> | ||
| + | |||
| + | [[File:T--NYMU-Taipei—Holin-Endolysin-REcheck.png|250px]] | ||
| + | |||
| + | |||
| + | |||
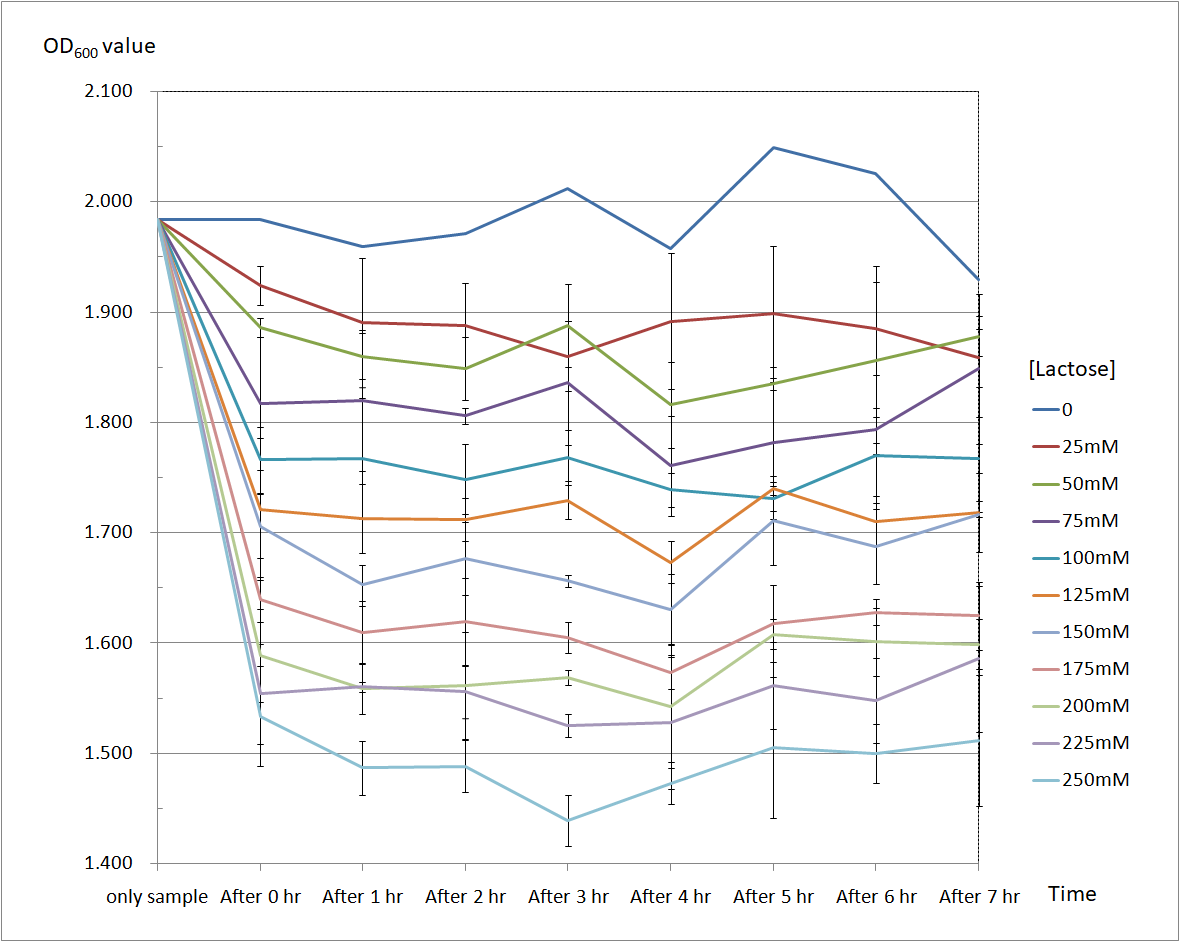
| + | Figure 6 and Figure 7 are the results of suicide mechanism functional test. | ||
| + | Both figure show that our suicide mechanism can be induced by adding lactose, and the effectiveness of suicide mechanism goes better as the concentration of lactose goes higher. | ||
| + | |||
| + | |||
| + | Figure 6 is the bacterium with holin-endolysin construct. | ||
| + | As figure 6 shows, the suicide mechanism is induced immediately when lactose is added into the samples. Besides, we can see that the lactose concentration and the OD value of the bacterium with holin-endolysin construct are positively correlated, which means our suicide mechanism does work. <br> | ||
| + | |||
| + | <b>Figure 6 </b> | ||
| + | |||
| + | [[File:BBa K2350020 LineGraph.png|600px]] | ||
| + | |||
| + | |||
| + | |||
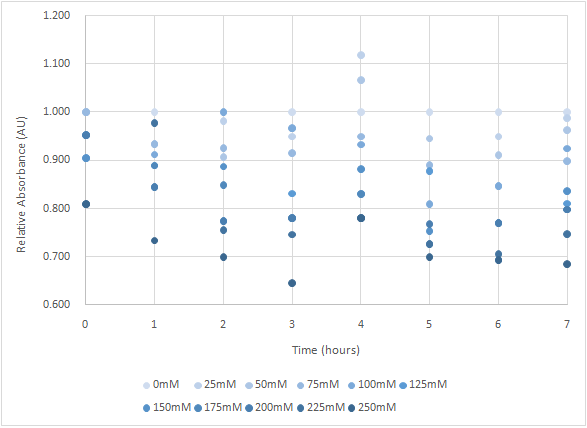
| + | Figure 7 is the bacterium with holin-endolysin-NrtA construct. | ||
| + | As figure 7 shows, the trend of the relative absorbance is downward as the lactose is added to induce the suicide mechanism. The concentration of lactose is also positively correlated with the declining degree of relative absorbance. <br> | ||
| + | |||
| + | <b>Figure 7 </b> | ||
| + | |||
| + | [[File:T--NYMU-Taipei--TAS-FunctionalTest.png|600px]] | ||
Latest revision as of 10:58, 30 October 2017
Holin from lambda phage

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Holin is a generic term to describe a group of small proteins produced by double-stranded DNA bacteriophage to trigger holes formation at the end of lytic cycle. In our project, we design our suicide switch based on the λ lysis model. The S holin, also called S105, encoded by S gene, a dual-start motif of λ phage, is an 105-amino-acid-residue CM protein with three transmembrane domains (TMD)[1]. S107, also called antiholin, is the other protein encoded by S gene, differing from the S holin only by the Met-Lys N-terminal extension. However, this difference confers to S107 an extra positive charge, which prevents its TMD1 from inserting into the CM[2]. Additionally, as its name suggests, S107 can bind to S105 and inhibit its function specifically[3]. In λ lysis system, S107 and S105 are encoded by S gene at ratio of approximately 1:2, which is defined by the two RNA structure, and if the amount of S107 is increased relative to S105, the 'lysis time' will be delayed[4]. The inhibition function of S107 can be subverted by collapsing proton motive force, which also allow insertion of TMD1 of S107 into CM, instantly increasing the amount of active holin by making previously inactive S107 - S105 complexes functional (Fig. 1).

Characterization
We choose λ lysis system to construct suicide switch due to its high efficiency and natural occurrence, and we introduce both endolysin and holin because of their cooperativity in cell lysis, which improves the performance of our suicide switch. In our design, endolysin is controlled by a constitutive promoter while holin by inducible promoter, Plac, because high concentration of holin can cause cell death alone (Fig. 4).

We transformed the two plasmids into E.coli. Then, 10mM of inducer was applied and the growth rate was measured. Compared with the bacteria carrying blank plasmid, the efficiency of our suicide switch can be evaluated.

The Fig. 5(a) shows that the difference between the OD595nm of experimental group and control group is obvious in the late logarithmic phase. The Fig. 5(b) shows that the growth curve of the E. coli carrying blank plasmid after the addition of 10mM IPTG is nearly coincident with that without addition of IPTG, excluding the possibility that the toxicity of IPTG leads to the noticeable OD595nm’s difference. Hence, the OD595nm’s difference should be caused by the slowed growth rate or cell death, and combining the working mechanism of holin and endolysin, we believe the cell death may be the main cause and our suicide switch may have some bactericidal effect.
In this experiment, every group did not enter the stationary phase and we thought better results could be attained by prolonging culture time. One possible reason for this performance of our suicide switch is that the expression of holin and endolysin is not enough to cause cell lysis. Hence, in order to test the performance of Plac, we will construct a plasmid where the expression of GFP is under control of Plac and measure the fluorescence intensity after the addition of a gradient of IPTG. According to the results of the experiment above, we will find an appropriate concentration of IPTG to get a better result if necessary. Besides, the relatively low toxicity of holin and endolysin may be another cause and we can choose the CcdA/CcdB Type II Toxin-antitoxin system instead in our future work because it have been proved that the CcdB, a topoisomerase poison targeting the GyrA subunit of DNA gyrase, shows strong toxicity to E. coli.
References
<p>[1]Gründling, A., Bläsi, U., & Young, R. (2000). Biochemical and genetic evidence for three transmembrane domains in the class I holin, lambda S. Journal of Biological Chemistry, 275(2), 769-776.[2]Young, R., Wang, I. N., & Roof, W. D. (2000). Phages will out: strategies of host cell lysis. Trends in microbiology, 8(3), 120-128.
[3]Bläsi, U., Chang, C. Y., Zagotta, M. T., Nam, K. B., & Young, R. (1990). The lethal lambda S gene encodes its own inhibitor. The EMBO journal, 9(4), 981.
[4]Bläsi, U., Nam, K., Hartz, D., Gold, L., & Young, R. (1989). Dual translational initiation sites control function of the lambda S gene. The EMBO journal, 8(11), 3501.
Team NYMU-Taipei 2017: Successfully construct suicide mechanism by T4 phage
Improvement
The gene sequences and the structures of holin in different species of bacteriophages are also different. In NYMU-Taipei 2017 team’s project, we choose the holin from enterobacteria phage T4. The T4 holin we used is an iGEM released part, BBa_K112000. We combine T4 holin with a lactose-induced promoter (BBa_R0010), a ribosome binding site (BBa_B0034) and a double terminator (BBa_B0010 and BBa_B0012). Therefore, holin can be induced by lactose and regulate the suicide mechanism. Moreover, we construct T4 holin and T4 endolysin together as a functional suicide mechanism. Besides, in our team NYMU-Taipei 2017 project, we also put the suicide mechanism into practice by constructing them with NrtA.
Result
Figure 1 is the gene map of T4 holin construct, pSB1C3-J04450-K112000.
Figure 1
Figure 2 is electrophoresis result of holin (from BBa_K112000) PCR product.
The marker is 100bp. The length is 657bp as expected.
Figure 2
Figure 3 is electrophoresis result of holin backbone (from BBa_J04450) PCR product. The marker is 1kb. The length is 2458bp as expected.
Figure 3
Figure 4 is the gene map of suicide mechanism construct, pSB1C3- T4 holin- T4 endolysin.
Figure 4
Figure 5 is restriction enzyme check electrophoresis result of T4 holin- T4 endolysin construct.
The marker is 1kb. We use XmaI to digest the sample. The total length is 3843bp as expected.
Figure 5
Figure 6 and Figure 7 are the results of suicide mechanism functional test. Both figure show that our suicide mechanism can be induced by adding lactose, and the effectiveness of suicide mechanism goes better as the concentration of lactose goes higher.
Figure 6 is the bacterium with holin-endolysin construct.
As figure 6 shows, the suicide mechanism is induced immediately when lactose is added into the samples. Besides, we can see that the lactose concentration and the OD value of the bacterium with holin-endolysin construct are positively correlated, which means our suicide mechanism does work.
Figure 6
Figure 7 is the bacterium with holin-endolysin-NrtA construct.
As figure 7 shows, the trend of the relative absorbance is downward as the lactose is added to induce the suicide mechanism. The concentration of lactose is also positively correlated with the declining degree of relative absorbance.
Figure 7