Difference between revisions of "Part:BBa K1351040"
| (8 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1351040 short</partinfo> | <partinfo>BBa_K1351040 short</partinfo> | ||
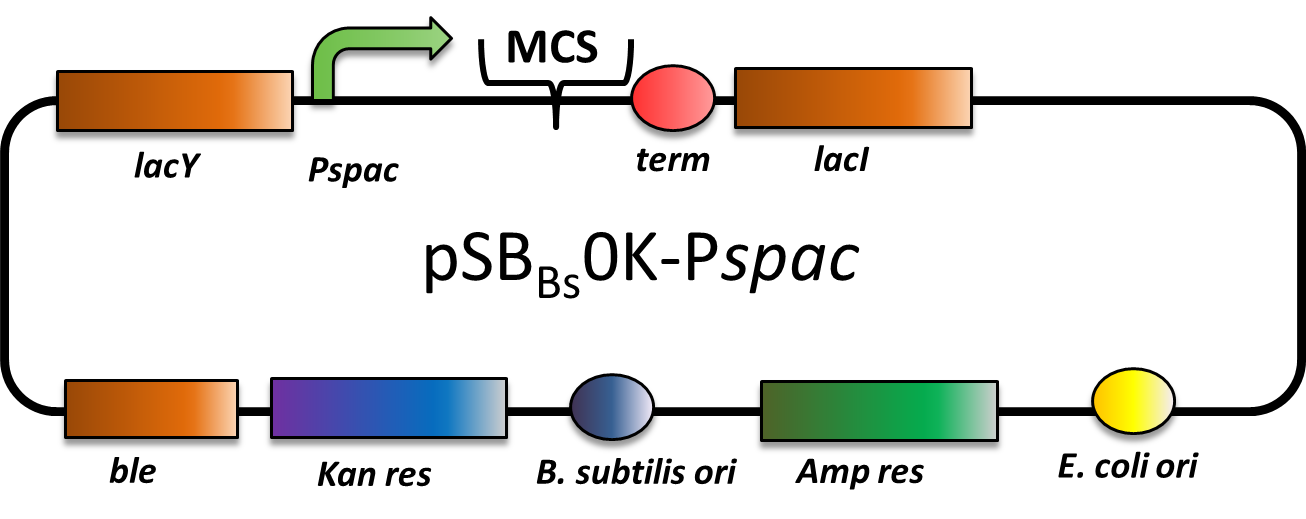
| − | pBS0K<i>Pspac</i>, an IPTG-inducible replicative expression vector for ''Bacillus subtilis''. | + | pBS0K<i>Pspac</i>, an IPTG-inducible replicative expression vector for ''Bacillus subtilis''. It is replicative both in ''E.coli'' and ''B.subtilis''. It has an ampicillin resistance for cloning in ''E.coli'' and kanamycin resistance for selection in ''B. subtilis''. The multiple cloning site is downstream of a P<sub>''spac''</sub> promoter, that is inducible with IPTG (Isopropyl-β-D-thiogalactopyranosid). |
| + | |||
| + | [[Image:LMU-Munich-PSBBs0K-Pspac.png|600px]] | ||
| + | |||
| + | <p align="justify"> | ||
| + | pSB<sub>''Bs''</sub>0K-P<sub><i>spac</i></sub> is a replicative expression vector with a kanamycin resistance. The IPTG (isopropylbeta-D-thiogalactopyranoside)-inducible Promoter P<sub>''spac''</sub> is followed by the multiple cloning site and a terminator. Also expressed are ''lac''Y, a transporter for IPTG (naturally allolactose), and ''lac'', the repressor. In presence of IPTG, LacI releases from the promoter and the gene of interest is expressed. | ||
| + | </p> | ||
| + | |||
| + | <p align="justify"> | ||
| + | The ''ble'' gene encodes the bleomycin resisitance protein (BRP) which can be selected for by bleomycine or phleomycin in ''E. coli'' and ''B. subtilis''. We did not use this resistance. | ||
| + | </p> | ||
| + | |||
| + | <p align="justify"> | ||
| + | The presence of the vector can be checked for via colony PCR with the primers: CTACATCCAGAACAACCTCTGC and TTCGGAAGGAAATGATGACCTC. | ||
| + | |||
| + | This backbone is a ''BioBricked'' version of the ''B. subtilis'' overexpression vector pDG148. Reference: [http://www.ncbi.nlm.nih.gov/pubmed/11728721 Joseph ''et al.''] | ||
| + | |||
| + | For handling of ''B. subtilis'' vectors, please see [https://static.igem.org/mediawiki/2012/c/c4/LMU-Munich_2012_Bacillus_subtilis_vectors.pdf here]. | ||
| + | |||
| + | The transformation into ''B. subtilis'' is explained [https://static.igem.org/mediawiki/2012/4/41/LMU-Munich_2012_Transformation_of_Bacillus_subtilis.pdf here]. | ||
| + | |||
| + | |||
| + | |||
| + | This is the "repaired" version of [https://parts.igem.org/Part:BBa_K823026 pSB<sub>Bs</sub>0K-P<sub>spac</sub>], where a single nucleotide loss of the lacI gene was reintroduced. | ||
| + | |||
| + | [[File:LMU14_GFP_pBS0K-Pspac1.jpg|250px|thumb|right|Fluorescence of GFP under expression of promoter P<sub>spac</sub>, induced with 1µM IPTG]] | ||
| + | |||
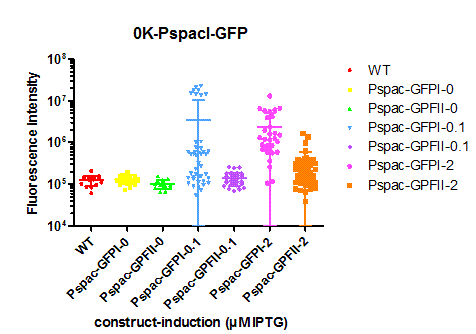
| + | We tested its expression by inserting [https://parts.igem.org/Part:BBa_K823039 ''gfp''] into the MCS downstream of P<sub>spac</sub> and transforming the plasmid into ''B. subtilis'' W168. Two independent clones were induced with 0, 0.1 or 2 µM IPTG and checked for fluorescence under the microscope. The fluorescence intensity of at least 100 cells was measured with imageJ. | ||
| + | |||
| + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[Image:LMU14_PBS0KPspac-activity.png|500px|center]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
| + | <font color="#000000"; size="2">''lacZ''-activity in Miller units in pSB<sub>Bs</sub>0K-P<sub>''spac''</sub> depending on ''B. subtilis'' growth phase.</font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | The vector is now IPTG-inducible, but high induction only leads to low and heterogenous gene expression. | ||
| + | |||
| + | |||
| + | This part is used in the 2014 LMU-Munich iGEM project [http://2014.igem.org/Team:LMU-Munich BaKillus]. | ||
| + | |||
| + | |||
| + | |||
| + | =<b>Note from iGEM Toulouse 2016:</b>= | ||
| + | This plasmid is the only replicative plasmid for <i>Bacillus subtilis</i> in the registry. As so, this BioBrick was improved by the iGEM Toulouse 2016 team by reducing its size down to 5827pb (compared to the 9358pb of the entire pSBBsOK -P). This makes clonings a lot more convenient. This new version is available at https://parts.igem.org/Part:BBa_K1937001. It is also described at http://2016.igem.org/Team:Toulouse_France/Description | ||
| + | |||
| + | |||
| + | Note that iGEM Toulouse 2016 also improved this backbone by creating from it a part that could turn any pSB1C3-based plasmid in a <i>E.coli/B.subtilis</i> shuttle vector. This part is named OriKan and can be found at https://parts.igem.org/Part:BBa_K1937002. | ||
| + | While <i>Bacillus subtilis</i> is of huge interest for a growing number of iGEM projects, it is not easy to develop new parts as it is required that they are registered after sub-cloning in the <i>E. coli</i> plasmid pSB1C3. In this context, the OriKAn part opens the whole iGEM registry to <i>Bacillus</i>-based projects. Its construction and validation are presented at http://2016.igem.org/Team:Toulouse_France/Description. | ||
| + | |||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 09:25, 15 October 2016
pBS0KPspac, an IPTG-inducible replicative expression vector for
pBS0KPspac, an IPTG-inducible replicative expression vector for Bacillus subtilis. It is replicative both in E.coli and B.subtilis. It has an ampicillin resistance for cloning in E.coli and kanamycin resistance for selection in B. subtilis. The multiple cloning site is downstream of a Pspac promoter, that is inducible with IPTG (Isopropyl-β-D-thiogalactopyranosid).
pSBBs0K-Pspac is a replicative expression vector with a kanamycin resistance. The IPTG (isopropylbeta-D-thiogalactopyranoside)-inducible Promoter Pspac is followed by the multiple cloning site and a terminator. Also expressed are lacY, a transporter for IPTG (naturally allolactose), and lac, the repressor. In presence of IPTG, LacI releases from the promoter and the gene of interest is expressed.
The ble gene encodes the bleomycin resisitance protein (BRP) which can be selected for by bleomycine or phleomycin in E. coli and B. subtilis. We did not use this resistance.
The presence of the vector can be checked for via colony PCR with the primers: CTACATCCAGAACAACCTCTGC and TTCGGAAGGAAATGATGACCTC. This backbone is a BioBricked version of the B. subtilis overexpression vector pDG148. Reference: [http://www.ncbi.nlm.nih.gov/pubmed/11728721 Joseph et al.] For handling of B. subtilis vectors, please see here. The transformation into B. subtilis is explained here. This is the "repaired" version of pSBBs0K-Pspac, where a single nucleotide loss of the lacI gene was reintroduced.
We tested its expression by inserting gfp into the MCS downstream of Pspac and transforming the plasmid into B. subtilis W168. Two independent clones were induced with 0, 0.1 or 2 µM IPTG and checked for fluorescence under the microscope. The fluorescence intensity of at least 100 cells was measured with imageJ.
|
The vector is now IPTG-inducible, but high induction only leads to low and heterogenous gene expression.
This part is used in the 2014 LMU-Munich iGEM project [http://2014.igem.org/Team:LMU-Munich BaKillus].
Note from iGEM Toulouse 2016:
This plasmid is the only replicative plasmid for Bacillus subtilis in the registry. As so, this BioBrick was improved by the iGEM Toulouse 2016 team by reducing its size down to 5827pb (compared to the 9358pb of the entire pSBBsOK -P). This makes clonings a lot more convenient. This new version is available at https://parts.igem.org/Part:BBa_K1937001. It is also described at http://2016.igem.org/Team:Toulouse_France/Description
Note that iGEM Toulouse 2016 also improved this backbone by creating from it a part that could turn any pSB1C3-based plasmid in a E.coli/B.subtilis shuttle vector. This part is named OriKan and can be found at https://parts.igem.org/Part:BBa_K1937002.
While Bacillus subtilis is of huge interest for a growing number of iGEM projects, it is not easy to develop new parts as it is required that they are registered after sub-cloning in the E. coli plasmid pSB1C3. In this context, the OriKAn part opens the whole iGEM registry to Bacillus-based projects. Its construction and validation are presented at http://2016.igem.org/Team:Toulouse_France/Description.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 8268
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NotI site found at 9
Illegal NotI site found at 8274 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 8268
Illegal BglII site found at 5862
Illegal BamHI site found at 1378 - 23INCOMPATIBLE WITH RFC[23]Illegal prefix found at 8268
Illegal suffix found at 2 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found at 8268
Plasmid lacks a suffix.
Illegal XbaI site found at 8283
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NgoMIV site found at 2925
Illegal AgeI site found at 5473
Illegal AgeI site found at 6435
Illegal AgeI site found at 7110 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal BsaI.rc site found at 2749
Illegal BsaI.rc site found at 4188
Illegal BsaI.rc site found at 6704
Illegal SapI site found at 1666
Illegal SapI.rc site found at 5686