Difference between revisions of "Part:BBa K1420003"
(→Experimental Results) |
(→Summary) |
||
| (18 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1420003 short</partinfo> | <partinfo>BBa_K1420003 short</partinfo> | ||
| + | |||
| + | ==Summary== | ||
| + | |||
| + | <p>'''•''' ''merP'' codes for the enzyme MerP is a periplasmic transporter that brings mercury into the cytoplasm of the cell.</p> | ||
| + | <p>'''•''' It is a non-essential transporter, due to similar activity of MerT brining Hg(II) into the cell.</p> | ||
| + | <p>'''•''' Zone of Inhibition tests conducted in the absence of Hg(II) reductase with mercury(II) chloride show the function of MerT and MerP in transporting Hg(II) into the cell..</p> | ||
==Overview== | ==Overview== | ||
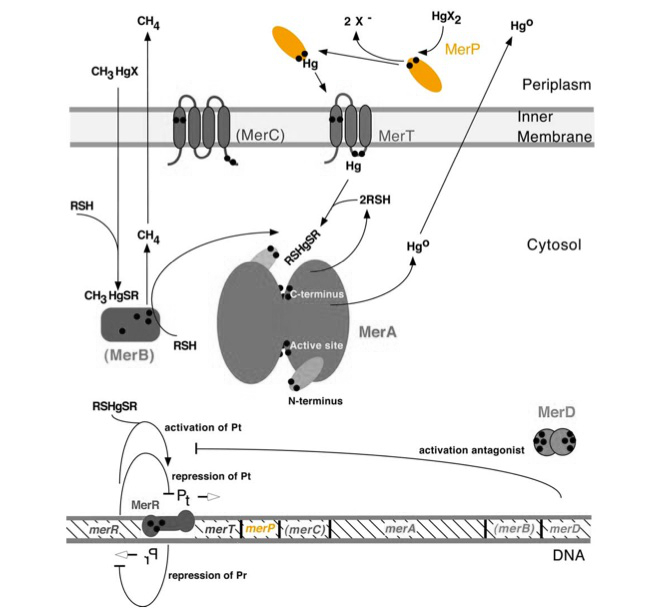
| − | <p>The gene ''merP'' (0.3Kb) encodes a periplasmic transport protein (MerP) that serves as part of the ''mer'' operon. MerP and the gene ''merP'' are highlighted in orange in Figure 1 below. The lower part of Figure 1 shows the arrangement of ''mer'' genes in the operon. ''merP'' is located downstream of ''merT'', another gene that encodes a transmembrane mercury transport protein.</p> | + | <p>The gene ''merP'' (0.3Kb) encodes a periplasmic transport protein (MerP) that serves as part of the ''mer'' operon. MerP and the gene ''merP'' are highlighted in orange in ''Figure 1'' below. The lower part of Figure 1 shows the arrangement of ''mer'' genes in the operon. ''merP'' is located downstream of ''merT'', another gene that encodes a transmembrane mercury transport protein.</p> |
[[File:MerP_gene.jpg|center|480px|x]] | [[File:MerP_gene.jpg|center|480px|x]] | ||
<p></p> | <p></p> | ||
| − | + | ''Figure 1.'' Model of mercury resistance operon. The symbol • indicates a cysteine residue. RSH indicates cytosolic thiol redox buffers such as glutathione. Figure 1 shows the interactions of MerP, in orange, with mercury compounds and other gene products of ''mer'' operon. (This figure is adapted from "Bacterial mercury resistance from atoms to ecosystems." <sup>1</sup>) | |
==Function== | ==Function== | ||
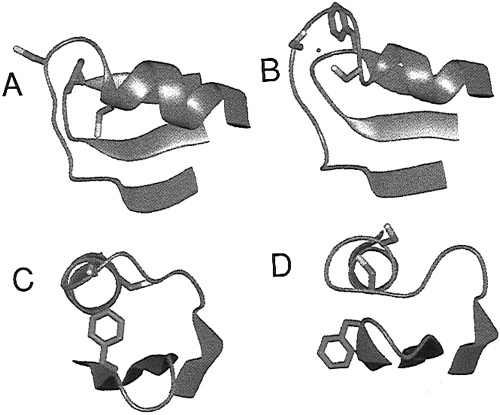
| − | MerP is a 72 amino acid periplasmic transport protein. MerP functions as a monomer with two alpha helices overlaying a four-strand antiparallel beta sheet. As shown in Figure 2, MerP binds a single Hg(II) ion at two conserved cysteine residues, these cysteines define the metal binding motif of MerP. Similar motifs are also found in other proteins that are involved in the transportation of thiophilic metal cations. The MerP cysteine residues take up a Hg(II) ion and remove any attached ligands before passing the ion to the MerT transmembrane protein. | + | MerP is a 72 amino acid periplasmic transport protein. MerP functions as a monomer with two alpha helices overlaying a four-strand antiparallel beta sheet. As shown in ''Figure 2'', MerP binds a single Hg(II) ion at two conserved cysteine residues, these cysteines define the metal binding motif of MerP. Similar motifs are also found in other proteins that are involved in the transportation of thiophilic metal cations. The MerP cysteine residues take up a Hg(II) ion and remove any attached ligands before passing the ion to the MerT transmembrane protein. |
[[File:MerP_structure.jpg|center]] | [[File:MerP_structure.jpg|center]] | ||
| − | + | ''Figure 2.'' Structure of MerP as determined by NMR studies. A shows a reduced MerP as viewed from above. B shows a MerP binding a Hg(II) ion, showing the two cysteine residues that become surface exposed. C and D show the protein as viewed down a helix. (Image source adapted from "Structures of the Reduced and Mercury-Bound Forms of MerP, the Periplasmic Protein from the Bacterial Mercury Detoxification System."<sup>2</sup>) | |
| Line 25: | Line 31: | ||
==Experimental Results== | ==Experimental Results== | ||
[[File:MNiGEM_ZOI_MerP.jpg|center|400px|x]] | [[File:MNiGEM_ZOI_MerP.jpg|center|400px|x]] | ||
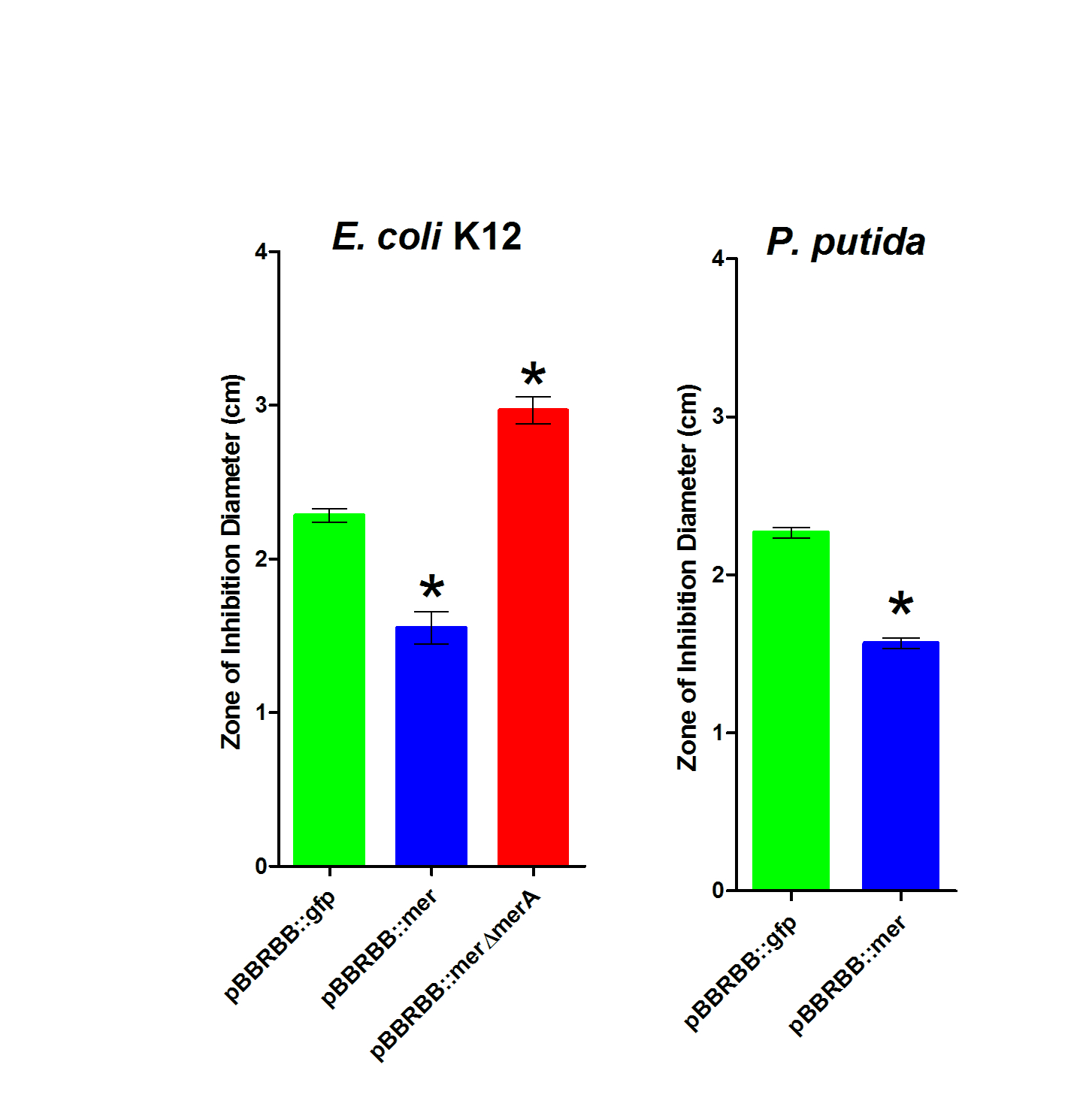
| − | Figure 3. Zone of Inhibition Plate Assasys. Strains were tested in triplicate and included ''E. coli'' K12 pBBRBB::''mer'', ''E. coli'' K12 pBBRBB::''gfp'', ''E. coli'' K12 pBBRBB::''merΔmerA'' (mer operon with ''merA'' deleted), ''P. putida'' pBBRBB::''mer'', and ''P. putida'' pBBRBB::''gfp''. | + | ''Figure 3.'' Zone of Inhibition Plate Assasys. Strains were tested in triplicate and included ''E. coli'' K12 pBBRBB::''mer'', ''E. coli'' K12 pBBRBB::''gfp'', ''E. coli'' K12 pBBRBB::''merΔmerA'' (mer operon with ''merA'' deleted), ''P. putida'' pBBRBB::''mer'', and ''P. putida'' pBBRBB::''gfp''. |
| − | To complete zone of inhibition plate assays, 10µL of a 0.1M HgCl<sub>2</sub> solution was spotted on a filter disk in the middle of an agar plate. Bacteria that are resistant to mercury are able to grow closer to the disc and hence have smaller zones of inhibition. On the other hand, bacteria that are sensitive to mercury are not able to grow near the filter disc resulting in a larger zone of inhibition and hence a larger diameter. As shown in Figure 3, for ''E. coli'', the strain harboring the ''mer'' operon (pBBRBB::''mer'') had the smallest zones of inhibition indicating enhanced resistance to Hg(II) as compared to the negative control (pBBRBB::''gfp''). In ''E. coli'' containing the ''mer'' operon with ''merA'' deleted (pBBRBB:''merΔmerA''), the zone of inhibition was larger than the negative control. This result is due to the fact that MerT and MerP facilitate mercury transport into cells, but without MerA, cells are unable to detoxify/reduce Hg(II) to Hg(0) leading to increased sensitivity. Increased sensitivity to Hg(II) with ''merT'' and ''merP'' in the absence of ''merA'' indicate that MerT and MerP are functioning together to transport Hg(II) into the cell. | + | To complete zone of inhibition plate assays, 10µL of a 0.1M HgCl<sub>2</sub> solution was spotted on a filter disk in the middle of an agar plate. Bacteria that are resistant to mercury are able to grow closer to the disc and hence have smaller zones of inhibition. On the other hand, bacteria that are sensitive to mercury are not able to grow near the filter disc resulting in a larger zone of inhibition and hence a larger diameter. As shown in ''Figure 3'', for ''E. coli'', the strain harboring the ''mer'' operon (pBBRBB::''mer'') had the smallest zones of inhibition indicating enhanced resistance to Hg(II) as compared to the negative control (pBBRBB::''gfp''). In ''E. coli'' containing the ''mer'' operon with ''merA'' deleted (pBBRBB:''merΔmerA''), the zone of inhibition was larger than the negative control. This result is due to the fact that MerT and MerP facilitate mercury transport into cells, but without MerA, cells are unable to detoxify/reduce Hg(II) to Hg(0) leading to increased sensitivity. Increased sensitivity to Hg(II) with ''merT'' and ''merP'' in the absence of ''merA'' indicate that MerT and MerP are functioning together to transport Hg(II) into the cell. |
| − | + | ||
| + | == References == | ||
| + | <p> 1. T. Barkay et al (2003). "Bacterial mercury resistance from atoms to ecosystems." ''FEMS Microbiology Reviews'' <b>27</b>: 355-384.</p> | ||
| + | <p></p> | ||
| + | <p>2. R. Steele et. al. (1997). "Structures of the Reduced and Mercury-Bound Forms of MerP, the Periplasmic Protein from the Bacterial Mercury Detoxification System". ''Biochemistry'' '''36'''(23): 6885–6895.</p> | ||
| + | <p></p> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| Line 37: | Line 47: | ||
<!-- --> | <!-- --> | ||
| − | + | '''Sequence and Features''' | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<partinfo>BBa_K1420003 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1420003 SequenceAndFeatures</partinfo> | ||
Latest revision as of 03:40, 18 October 2014
MerP, mercuric transport protein periplasmic component
Summary
• merP codes for the enzyme MerP is a periplasmic transporter that brings mercury into the cytoplasm of the cell.
• It is a non-essential transporter, due to similar activity of MerT brining Hg(II) into the cell.
• Zone of Inhibition tests conducted in the absence of Hg(II) reductase with mercury(II) chloride show the function of MerT and MerP in transporting Hg(II) into the cell..
Overview
The gene merP (0.3Kb) encodes a periplasmic transport protein (MerP) that serves as part of the mer operon. MerP and the gene merP are highlighted in orange in Figure 1 below. The lower part of Figure 1 shows the arrangement of mer genes in the operon. merP is located downstream of merT, another gene that encodes a transmembrane mercury transport protein.
Figure 1. Model of mercury resistance operon. The symbol • indicates a cysteine residue. RSH indicates cytosolic thiol redox buffers such as glutathione. Figure 1 shows the interactions of MerP, in orange, with mercury compounds and other gene products of mer operon. (This figure is adapted from "Bacterial mercury resistance from atoms to ecosystems." 1)
Function
MerP is a 72 amino acid periplasmic transport protein. MerP functions as a monomer with two alpha helices overlaying a four-strand antiparallel beta sheet. As shown in Figure 2, MerP binds a single Hg(II) ion at two conserved cysteine residues, these cysteines define the metal binding motif of MerP. Similar motifs are also found in other proteins that are involved in the transportation of thiophilic metal cations. The MerP cysteine residues take up a Hg(II) ion and remove any attached ligands before passing the ion to the MerT transmembrane protein.
Figure 2. Structure of MerP as determined by NMR studies. A shows a reduced MerP as viewed from above. B shows a MerP binding a Hg(II) ion, showing the two cysteine residues that become surface exposed. C and D show the protein as viewed down a helix. (Image source adapted from "Structures of the Reduced and Mercury-Bound Forms of MerP, the Periplasmic Protein from the Bacterial Mercury Detoxification System."2)
MerP is not essential for Hg(II) uptake, as MerT can import Hg(II) by itself. Additionally, there is the possibility of a Cys14Ser mutation on MerP which blocks Hg(II) uptake by MerT. If merP is expressed alone, it will exist in an oxidized state, which is less stable than the reduced state it is found in when expressed as part of the greater mer operon. In the periplasm, MerP competes with thiol-mediated redox proteins for Hg(II). Like periplasmic nutrient transporters, MerP is the most abundantly synthesized protein in its respective operon due to its role in periplasmic Hg(II) transport.
Experimental Results
Figure 3. Zone of Inhibition Plate Assasys. Strains were tested in triplicate and included E. coli K12 pBBRBB::mer, E. coli K12 pBBRBB::gfp, E. coli K12 pBBRBB::merΔmerA (mer operon with merA deleted), P. putida pBBRBB::mer, and P. putida pBBRBB::gfp.
To complete zone of inhibition plate assays, 10µL of a 0.1M HgCl2 solution was spotted on a filter disk in the middle of an agar plate. Bacteria that are resistant to mercury are able to grow closer to the disc and hence have smaller zones of inhibition. On the other hand, bacteria that are sensitive to mercury are not able to grow near the filter disc resulting in a larger zone of inhibition and hence a larger diameter. As shown in Figure 3, for E. coli, the strain harboring the mer operon (pBBRBB::mer) had the smallest zones of inhibition indicating enhanced resistance to Hg(II) as compared to the negative control (pBBRBB::gfp). In E. coli containing the mer operon with merA deleted (pBBRBB:merΔmerA), the zone of inhibition was larger than the negative control. This result is due to the fact that MerT and MerP facilitate mercury transport into cells, but without MerA, cells are unable to detoxify/reduce Hg(II) to Hg(0) leading to increased sensitivity. Increased sensitivity to Hg(II) with merT and merP in the absence of merA indicate that MerT and MerP are functioning together to transport Hg(II) into the cell.
References
1. T. Barkay et al (2003). "Bacterial mercury resistance from atoms to ecosystems." FEMS Microbiology Reviews 27: 355-384.
2. R. Steele et. al. (1997). "Structures of the Reduced and Mercury-Bound Forms of MerP, the Periplasmic Protein from the Bacterial Mercury Detoxification System". Biochemistry 36(23): 6885–6895.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]