Difference between revisions of "Part:BBa K1321370"
| Line 8: | Line 8: | ||
Note that the start and stop codon, plus 6 bp either side of the sequence, are included the RFC25 prefix and suffix which is not shown. | Note that the start and stop codon, plus 6 bp either side of the sequence, are included the RFC25 prefix and suffix which is not shown. | ||
| + | |||
| + | For reference the cellulose binding domain binding capability to bacterial cellulose was measured relative to other cellulose binding domains when fused to sfGFP, the data for which can be seen [https://parts.igem.org/Part:BBa_K1321371 here] - K1321371. | ||
Latest revision as of 05:21, 2 November 2014
NiBP + Linker-CBDcipA-Linker
**Important note**: Please be aware we discovered that this part is out of frame, leading to an incorrect translation and read-through of the stop-codon. This also means any subsequent fusion proteins to the C-terminal are out of frame. This is because there is a deletion of a base compared to the equivalent coding sequence from Heliobacter pylori (GenBank CP000012.1 bp1391851-1392033, reverse complement strand). This existed already in BBa_K1151001 from which we cloned our part. We hope that this part will still be useful, since the error could be corrected relatively easily by a team who wishes to it by site-directed-mutagenesis PCR to insert the missing nucleotide.
This construct is part of a library of fusions with cellulose binding domains which we designed to bind to cellulose and enable capture of heavy metals. Other fusion parts with this metal binding protein can be seen in the table below: 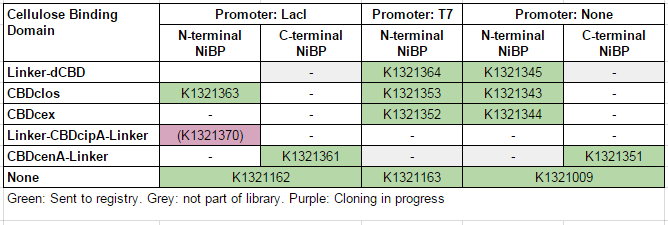
Note that the start and stop codon, plus 6 bp either side of the sequence, are included the RFC25 prefix and suffix which is not shown.
For reference the cellulose binding domain binding capability to bacterial cellulose was measured relative to other cellulose binding domains when fused to sfGFP, the data for which can be seen here - K1321371.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 330
- 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 330
- 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 330
- 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 330
- 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 330
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 174
