Difference between revisions of "Part:BBa K1420005"
| (33 intermediate revisions by 5 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1420005 short</partinfo> | <partinfo>BBa_K1420005 short</partinfo> | ||
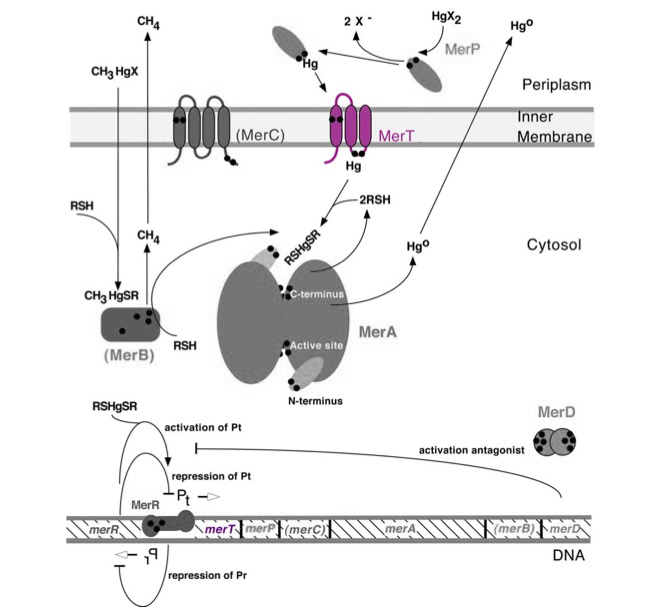
| − | <p>Transmembrane mercuric binding gene ''merT'' (0.3Kb) encodes MerT, which transports Hg(II) species from the periplasm through the membrane. MerT and gene ''merT'' are highlighted in purple in Figure 1. The lower part of the Figure 1 shows the arrangement of ''mer'' genes in the operon, and ''merT'' is located upstream of ''merP'', another gene that encodes a mercury transport protein.</p> | + | ==Summary== |
| + | <p>'''•''' MerT is a 116 kDa transmembrane protein that assists in transporting Hg(II) into the cytoplasm of the bacterial cell. </p> | ||
| + | <p>'''•''' Accomplishes transport of Hg(II) by interactions of cysteine residues along the protein and folding into the lowest possible energy structure. </p> | ||
| + | <p>'''•''' Zone of Inhibition tests conducted in the absence of Hg(II) reductase with mercury(II) chloride show the function of MerT and MerP in transporting Hg(II) into the cell.</p> | ||
| + | |||
| + | ==Overview== | ||
| + | <p>Transmembrane mercuric binding gene ''merT'' (0.3Kb) encodes MerT, which transports Hg(II) species from the periplasm through the membrane. MerT and gene ''merT'' are highlighted in purple in ''Figure 1''. The lower part of the ''Figure 1'' shows the arrangement of ''mer'' genes in the operon, and ''merT'' is located upstream of ''merP'', another gene that encodes a mercury transport protein. The gene ''merT'' is found in the ''Serratia marcescens'' plasmid pDU1358 downstream of the bidirectional promoter of ''merR,'' and it is 351 base pairs long.</p> | ||
[[File:MerT_gene.jpg|center|480px|x]] | [[File:MerT_gene.jpg|center|480px|x]] | ||
<p></p> | <p></p> | ||
| − | + | ''Figure 1.'' Model of transmembrane mercuric binding protein MerT. The symbol • indicates a cysteine residue. RSH indicates cytosolic thiol redox buffers such as glutathione. Figure 1 shows the interactions of MerT, in purple, with mercury compounds and other gene products of ''mer'' operon. (This figure is adapted from "Bacterial mercury resistance from atoms to ecosystems."<sup>1</sup> ) | |
<p></p> | <p></p> | ||
| − | + | ==Structure== | |
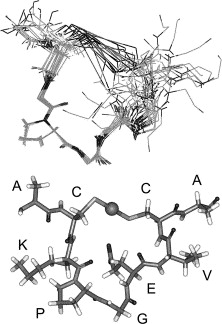
| + | MerT is a 116-residue (12.4 kDa) transmembrane protein. The exact structure is not known, but the protein is predicted to have three transmembrane helices with a cysteine pair accessible from the periplasmic side and another pair on the cytoplasmic side. A proposed representation of MerT bound to Hg(II) can be seen in ''Figure 2.'' | ||
| − | |||
| − | MerT | + | [[File:MerT image2.jpg|center|300px|]] |
| + | ''Figure 2.'' Structure of MerT bound to Hg(II). Top Panel: Images of the 20 lowest energy states that have been superimposed upon each other. Bottom Panel: Lowest energy structure of MerT in which the sphere represents the mercury. (Image source adapted from "Is the cytoplasmic loop of MerT, the mercuric ion transport protein, involved in mercury transfer to the mercuric reductase?." <sup>2</sup>) | ||
| − | + | ==Molecular Mechanism== | |
| − | + | ||
| − | + | ||
The transport mechanism of ionic mercury from the bacterial cell periplasm to cytosplasm is not completely understood. Hg(II) is transferred from the periplasmic cysteine pair on the first transmembrane helix to the cytoplasmic loop cysteine, where it is finally transferred to a cysteine pair at the N-terminus of the protein. The Hg(II) may go directly to MerA from the N-terminus of MerT or transfer to cytoplasmic low-molecular mass thiols. | The transport mechanism of ionic mercury from the bacterial cell periplasm to cytosplasm is not completely understood. Hg(II) is transferred from the periplasmic cysteine pair on the first transmembrane helix to the cytoplasmic loop cysteine, where it is finally transferred to a cysteine pair at the N-terminus of the protein. The Hg(II) may go directly to MerA from the N-terminus of MerT or transfer to cytoplasmic low-molecular mass thiols. | ||
| + | |||
| + | == Experimental Results == | ||
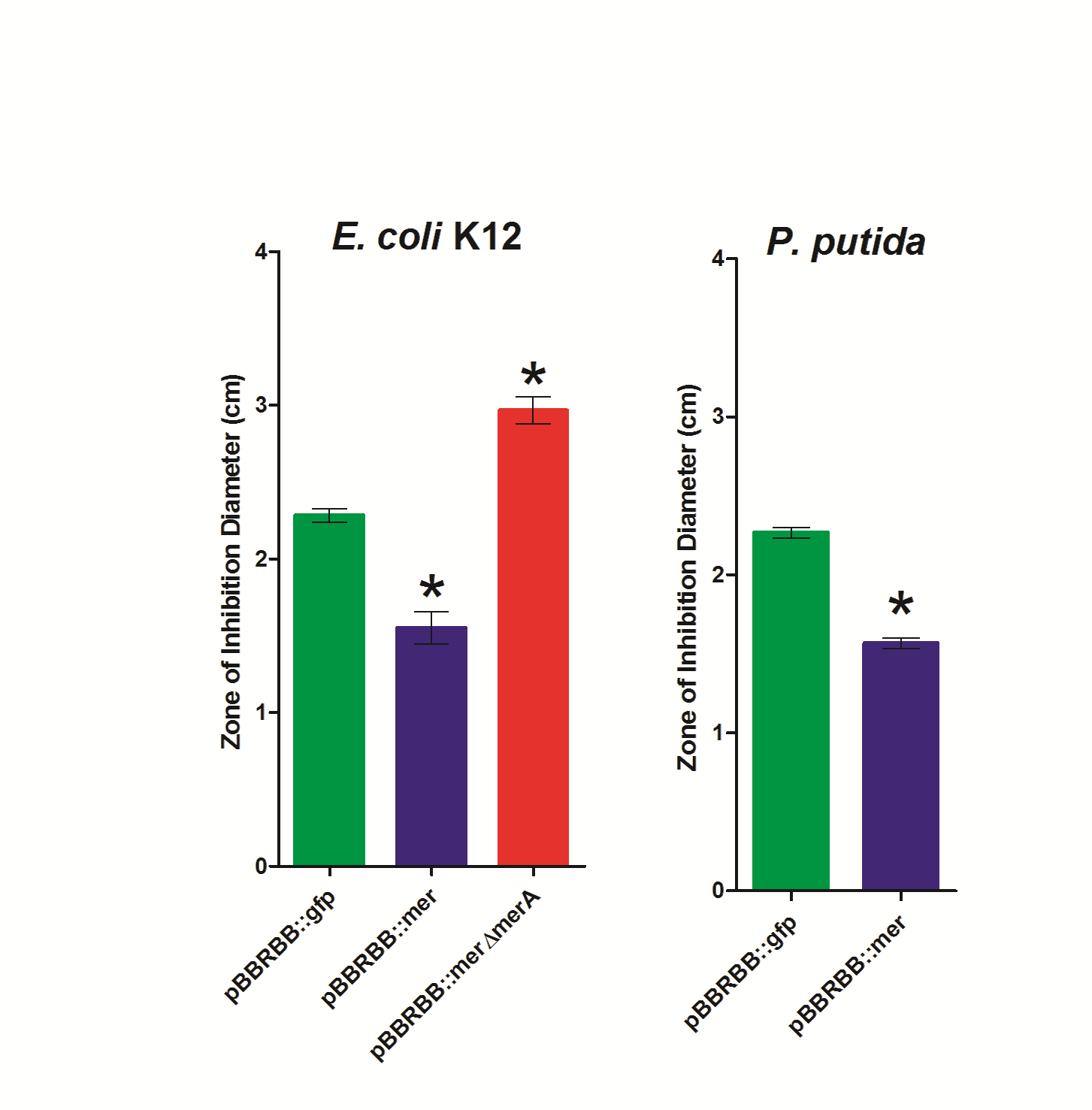
| + | [[File:2nd_zone_of_inhibition_data.png|center|400px|]] | ||
| + | ''Figure 3.'' Zone of Inhibition Plate Assasys. Strains were tested in triplicate and included ''E. coli'' K12 pBBRBB::''mer'', ''E. coli'' K12 pBBRBB::''gfp'', ''E. coli'' K12 pBBRBB::''merΔmerA'' (mer operon with ''merA'' deleted), ''P. putida'' pBBRBB::''mer'', and ''P. putida'' pBBRBB::''gfp''. | ||
| + | |||
| + | |||
| + | <p>To complete zone of inhibition plate assays, 10µL of a 0.1M HgCl<sub>2</sub> solution was spotted on a filter disk in the middle of an agar plate. Bacteria that are resistant to mercury are able to grow closer to the disc and hence have smaller zones of inhibition. On the other hand, bacteria that are sensitive to mercury are not able to grow near the filter disc resulting in a larger zone of inhibition and hence a larger diameter. As shown in Figure 3, for ''E. coli'', the strain harboring the ''mer'' operon (pBBRBB::''mer'') had the smallest zones of inhibition indicating enhanced resistance to Hg(II) as compared to the negative control (pBBRBB::''gfp''). In ''E. coli'' containing the ''mer'' operon with ''merA'' deleted (pBBRBB:''merΔmerA''), the zone of inhibition was larger than the negative control. This result is due to the fact that MerT and MerP facilitate mercury transport into cells, but without MerA, cells are unable to detoxify/reduce Hg(II) to Hg(0) leading to increased sensitivity. Increased sensitivity to Hg(II) with ''merT'' and ''merP'' in the absence of ''merA'' indicate that MerT and MerP are functioning together to transport Hg(II) into the cell. </p> | ||
| + | |||
| + | ==MIT_MAHE 2020== | ||
| + | '''Role of cysteine''' | ||
| + | |||
| + | The cysteine residues of merT play an important role in imparting mercury resistance shown by site directed mutagenesis (Morby et al. 1995). Mutations in these residues led to the decrease of mercury hypersensitivity and resistance (Hobman J.L. et al., 1996). A bacteria two-hybrid protein interaction assay showed that the interaction between the N-terminal region of mercuric reductase and cytoplasmic loop of mercuric ion transporter MerT was not enhanced by mercuric ion presence and occurs in vivo. | ||
| + | |||
| + | This interaction, however, did not require the cysteine residues in the respective interacting domains. Another assay study showed that the cysteines located in the cytoplasmic loop of MerT were essential for maximum rate of mercury volatilization. However, there was volatilization of mercury in the absence of these residues is also possible. These results first demonstrated the MerT-MerA interactions suggesting that for Hg2+ transfer from MerT to catalytic site of MerA occurred via the MerP-like N terminal domain and the interactions although not essential increases the system’s mercury detoxification efficiency (Schue M. et al., 2007). | ||
| + | |||
| + | |||
| + | |||
| + | '''Usage and biology''' | ||
| + | |||
| + | A study showed that cells expressing MerT or MerC had increased sensitivity to Hg(II) and C6H5Hg(I) than MerE and MerF (Sone Y. et al., 2013). | ||
| + | |||
| + | The location of MerP and MerT in the operon are close to each other suggesting that they work co-operatively. In another study, if MerA is not available to the cells, expression of merP and merT can possibly increase the sensitivity of the cells to Hg2+ (Deng and Wilson, 2001). | ||
| + | |||
| + | In a recent study involving marine mercury resistant strains, it has been shown that strains expressing only MerP and MerT have higher Hg2+ resistance than strains having MerP MerT and MerA showing that the contributions of these genes to mercury resistance is not cumulative. The possible reason for this could be that the burden of a greater number of proteins which reduces the efficiency of the proteins. It also showed that the Minimum inhibitory concentration of strains (marine) containing MerP and MerT is three times higher than strains containing only MerA suggesting that expression of MerP MerT imparts the bacteria with the ability to tolerate Hg2+ (Zhang J. et al., 2020). | ||
| + | |||
| + | == References == | ||
| + | <p>1. T. Barkay et al (2003). "Bacterial mercury resistance from atoms to ecosystems." ''FEMS Microbiology Reviews'' <b>27</b>: 355-384.</p> | ||
| + | <p></p> | ||
| + | <p>2. Rossy E et al. (2004). "Is the cytoplasmic loop of MerT, the mercuric ion transport protein, involved in mercury transfer to the mercuric reductase?" ''FEBS Lett.'' '''575''':86–90 </p> | ||
| + | |||
| + | <p>3. Deng, X., Wilson, D., 2001. Bioaccumulation of mercury from wastewater by genetically engineered Escherichia coli. Appl. Microbiol. Biot. 56, 276–279. </p> | ||
| + | |||
| + | <p>4. Hobman, J. L., & Brown, N. L. (1996). Overexpression of MerT, the mercuric ion transport protein of transposon Tn501, and genetic selection of mercury hypersensitivity mutations. Molecular & general genetics : MGG, 250(1), 129–134. https://doi.org/10.1007/BF02191833 </p> | ||
| + | |||
| + | <p>5. Schue, M., Glendinning, K. J., Hobman, J. L., & Brown, N. L. (2008). Evidence for direct interactions between the mercuric ion transporter (MerT) and mercuric reductase (MerA) from the Tn501 mer operon. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine, 21(2), 107–116. https://doi.org/10.1007/s10534-007-9097-4 </p> | ||
| + | |||
| + | <p>6. Sone, Y., Nakamura, R., Pan-Hou, H., Itoh, T., & Kiyono, M. (2013). Role of MerC, MerE, MerF, MerT, and/or MerP in resistance to mercurials and the transport of mercurials in Escherichia coli. Biological & pharmaceutical bulletin, 36(11), 1835–1841. https://doi.org/10.1248/bpb.b13-00554 </p> | ||
| + | |||
| + | <p>7. Zhang, J. , Zenga, Y., Liu, B., Deng, X., 2020. MerP/MerT-mediated mechanism: A different approach to mercury resistance and bioaccumulation by marine bacteria. Journal of hazardous material 388 (2020) 122022 </p> | ||
| + | <p></p> | ||
| + | |||
| + | '''Sequence and Features''' | ||
<!-- --> | <!-- --> | ||
| − | |||
<partinfo>BBa_K1420005 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1420005 SequenceAndFeatures</partinfo> | ||
| + | <!-- --> | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 10:32, 20 October 2020
merT, mercuric transport protein
Summary
• MerT is a 116 kDa transmembrane protein that assists in transporting Hg(II) into the cytoplasm of the bacterial cell.
• Accomplishes transport of Hg(II) by interactions of cysteine residues along the protein and folding into the lowest possible energy structure.
• Zone of Inhibition tests conducted in the absence of Hg(II) reductase with mercury(II) chloride show the function of MerT and MerP in transporting Hg(II) into the cell.
Overview
Transmembrane mercuric binding gene merT (0.3Kb) encodes MerT, which transports Hg(II) species from the periplasm through the membrane. MerT and gene merT are highlighted in purple in Figure 1. The lower part of the Figure 1 shows the arrangement of mer genes in the operon, and merT is located upstream of merP, another gene that encodes a mercury transport protein. The gene merT is found in the Serratia marcescens plasmid pDU1358 downstream of the bidirectional promoter of merR, and it is 351 base pairs long.
Figure 1. Model of transmembrane mercuric binding protein MerT. The symbol • indicates a cysteine residue. RSH indicates cytosolic thiol redox buffers such as glutathione. Figure 1 shows the interactions of MerT, in purple, with mercury compounds and other gene products of mer operon. (This figure is adapted from "Bacterial mercury resistance from atoms to ecosystems."1 )
Structure
MerT is a 116-residue (12.4 kDa) transmembrane protein. The exact structure is not known, but the protein is predicted to have three transmembrane helices with a cysteine pair accessible from the periplasmic side and another pair on the cytoplasmic side. A proposed representation of MerT bound to Hg(II) can be seen in Figure 2.
Figure 2. Structure of MerT bound to Hg(II). Top Panel: Images of the 20 lowest energy states that have been superimposed upon each other. Bottom Panel: Lowest energy structure of MerT in which the sphere represents the mercury. (Image source adapted from "Is the cytoplasmic loop of MerT, the mercuric ion transport protein, involved in mercury transfer to the mercuric reductase?." 2)
Molecular Mechanism
The transport mechanism of ionic mercury from the bacterial cell periplasm to cytosplasm is not completely understood. Hg(II) is transferred from the periplasmic cysteine pair on the first transmembrane helix to the cytoplasmic loop cysteine, where it is finally transferred to a cysteine pair at the N-terminus of the protein. The Hg(II) may go directly to MerA from the N-terminus of MerT or transfer to cytoplasmic low-molecular mass thiols.
Experimental Results
Figure 3. Zone of Inhibition Plate Assasys. Strains were tested in triplicate and included E. coli K12 pBBRBB::mer, E. coli K12 pBBRBB::gfp, E. coli K12 pBBRBB::merΔmerA (mer operon with merA deleted), P. putida pBBRBB::mer, and P. putida pBBRBB::gfp.
To complete zone of inhibition plate assays, 10µL of a 0.1M HgCl2 solution was spotted on a filter disk in the middle of an agar plate. Bacteria that are resistant to mercury are able to grow closer to the disc and hence have smaller zones of inhibition. On the other hand, bacteria that are sensitive to mercury are not able to grow near the filter disc resulting in a larger zone of inhibition and hence a larger diameter. As shown in Figure 3, for E. coli, the strain harboring the mer operon (pBBRBB::mer) had the smallest zones of inhibition indicating enhanced resistance to Hg(II) as compared to the negative control (pBBRBB::gfp). In E. coli containing the mer operon with merA deleted (pBBRBB:merΔmerA), the zone of inhibition was larger than the negative control. This result is due to the fact that MerT and MerP facilitate mercury transport into cells, but without MerA, cells are unable to detoxify/reduce Hg(II) to Hg(0) leading to increased sensitivity. Increased sensitivity to Hg(II) with merT and merP in the absence of merA indicate that MerT and MerP are functioning together to transport Hg(II) into the cell.
MIT_MAHE 2020
Role of cysteine
The cysteine residues of merT play an important role in imparting mercury resistance shown by site directed mutagenesis (Morby et al. 1995). Mutations in these residues led to the decrease of mercury hypersensitivity and resistance (Hobman J.L. et al., 1996). A bacteria two-hybrid protein interaction assay showed that the interaction between the N-terminal region of mercuric reductase and cytoplasmic loop of mercuric ion transporter MerT was not enhanced by mercuric ion presence and occurs in vivo.
This interaction, however, did not require the cysteine residues in the respective interacting domains. Another assay study showed that the cysteines located in the cytoplasmic loop of MerT were essential for maximum rate of mercury volatilization. However, there was volatilization of mercury in the absence of these residues is also possible. These results first demonstrated the MerT-MerA interactions suggesting that for Hg2+ transfer from MerT to catalytic site of MerA occurred via the MerP-like N terminal domain and the interactions although not essential increases the system’s mercury detoxification efficiency (Schue M. et al., 2007).
Usage and biology
A study showed that cells expressing MerT or MerC had increased sensitivity to Hg(II) and C6H5Hg(I) than MerE and MerF (Sone Y. et al., 2013).
The location of MerP and MerT in the operon are close to each other suggesting that they work co-operatively. In another study, if MerA is not available to the cells, expression of merP and merT can possibly increase the sensitivity of the cells to Hg2+ (Deng and Wilson, 2001).
In a recent study involving marine mercury resistant strains, it has been shown that strains expressing only MerP and MerT have higher Hg2+ resistance than strains having MerP MerT and MerA showing that the contributions of these genes to mercury resistance is not cumulative. The possible reason for this could be that the burden of a greater number of proteins which reduces the efficiency of the proteins. It also showed that the Minimum inhibitory concentration of strains (marine) containing MerP and MerT is three times higher than strains containing only MerA suggesting that expression of MerP MerT imparts the bacteria with the ability to tolerate Hg2+ (Zhang J. et al., 2020).
References
1. T. Barkay et al (2003). "Bacterial mercury resistance from atoms to ecosystems." FEMS Microbiology Reviews 27: 355-384.
2. Rossy E et al. (2004). "Is the cytoplasmic loop of MerT, the mercuric ion transport protein, involved in mercury transfer to the mercuric reductase?" FEBS Lett. 575:86–90
3. Deng, X., Wilson, D., 2001. Bioaccumulation of mercury from wastewater by genetically engineered Escherichia coli. Appl. Microbiol. Biot. 56, 276–279.
4. Hobman, J. L., & Brown, N. L. (1996). Overexpression of MerT, the mercuric ion transport protein of transposon Tn501, and genetic selection of mercury hypersensitivity mutations. Molecular & general genetics : MGG, 250(1), 129–134. https://doi.org/10.1007/BF02191833
5. Schue, M., Glendinning, K. J., Hobman, J. L., & Brown, N. L. (2008). Evidence for direct interactions between the mercuric ion transporter (MerT) and mercuric reductase (MerA) from the Tn501 mer operon. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine, 21(2), 107–116. https://doi.org/10.1007/s10534-007-9097-4
6. Sone, Y., Nakamura, R., Pan-Hou, H., Itoh, T., & Kiyono, M. (2013). Role of MerC, MerE, MerF, MerT, and/or MerP in resistance to mercurials and the transport of mercurials in Escherichia coli. Biological & pharmaceutical bulletin, 36(11), 1835–1841. https://doi.org/10.1248/bpb.b13-00554
7. Zhang, J. , Zenga, Y., Liu, B., Deng, X., 2020. MerP/MerT-mediated mechanism: A different approach to mercury resistance and bioaccumulation by marine bacteria. Journal of hazardous material 388 (2020) 122022
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 45
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 30