Difference between revisions of "Part:BBa K1326001"
Leah.Simmons (Talk | contribs) |
Leah.Simmons (Talk | contribs) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1326001 short</partinfo> | <partinfo>BBa_K1326001 short</partinfo> | ||
| − | Mg protoporphyrin IX S-adenosyl methionine O-methyl transferase - Magnesium-protoporphyrin O-methyltransferase (ChlM) [PMID: 12828371; 12489983; 4436384]; ChloroP 1.1 predicts cp location | + | Mg protoporphyrin IX S-adenosyl methionine O-methyl transferase - Magnesium-protoporphyrin O-methyltransferase (ChlM) [PMID: [http://www.ncbi.nlm.nih.gov/pubmed/12828371 12828371]; [http://www.ncbi.nlm.nih.gov/pubmed/12489983 12489983]; [http://www.ncbi.nlm.nih.gov/pubmed/4436384 4436384]]; ChloroP 1.1 predicts cp location |
<br><br> | <br><br> | ||
===Operon Usage=== | ===Operon Usage=== | ||
| Line 25: | Line 25: | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K1326001 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1326001 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <b> Amino Acid Sequence </b> | ||
| + | |||
| + | <FONT FACE="courier">MASEIAQTAD VGSLTFAVGG VGAVVGLGAL LVATDHQKRR SEQMKSFDGD EKEAVKDYFN<br> TAGFERWRKI YGETDEVNKV QLDIRTGHAQ TVDKVLRWVD EEGSVQGITV ADCGCGTGSL<br> AIQLALRGAA VSASDISAAM ASEAEQRYQQ AVAAGQGKAP KVAPKFEALD LESVKGKYDT<br> VTCLDVMIHY PQDKVDAMIT HLAGLSDRRL IISFAPKTLS YSILKRIGEL FPGPSKATRA<br> YLHREEDVEA ALKRAGFKVT KREMTATSFY FSRLLEAIRE </FONT> | ||
| + | |||
| + | References and documentation are available. | ||
| + | Please note the modified algorithm for extinction coefficient. | ||
| + | |||
| + | -------------------------------------------------------------------------------- | ||
| + | Number of amino acids: 280 | ||
| + | |||
| + | Molecular weight: 30440.6 | ||
| + | |||
| + | Theoretical pI: 6.25 | ||
| + | |||
| + | Amino acid composition: | ||
| + | <br> | ||
| + | Ala (A) 36 12.9%<br> | ||
| + | Arg (R) 17 6.1%<br> | ||
| + | Asn (N) 2 0.7%<br> | ||
| + | Asp (D) 18 6.4%<br> | ||
| + | Cys (C) 3 1.1%<br> | ||
| + | Gln (Q) 12 4.3%<br> | ||
| + | Glu (E) 20 7.1%<br> | ||
| + | Gly (G) 23 8.2%<br> | ||
| + | His (H) 5 1.8%<br> | ||
| + | Ile (I) 13 4.6%<br> | ||
| + | Leu (L) 22 7.9%<br> | ||
| + | Lys (K) 19 6.8%<br> | ||
| + | Met (M) 6 2.1%<br> | ||
| + | Phe (F) 10 3.6%<br> | ||
| + | Pro (P) 6 2.1%<br> | ||
| + | Ser (S) 18 6.4%<br> | ||
| + | Thr (T) 17 6.1%<br> | ||
| + | Trp (W) 2 0.7%<br> | ||
| + | Tyr (Y) 8 2.9%<br> | ||
| + | Val (V) 23 8.2%<br> | ||
| + | Pyl (O) 0 0.0%<br> | ||
| + | Sec (U) 0 0.0%<br> | ||
| + | |||
| + | (B) 0 0.0% | ||
| + | (Z) 0 0.0% | ||
| + | (X) 0 0.0% | ||
| + | |||
| + | |||
| + | Total number of negatively charged residues (Asp + Glu): 38 | ||
| + | Total number of positively charged residues (Arg + Lys): 36 | ||
| + | |||
| + | Atomic composition: | ||
| + | |||
| + | Carbon C 1340 | ||
| + | Hydrogen H 2150 | ||
| + | Nitrogen N 376 | ||
| + | Oxygen O 414 | ||
| + | Sulfur S 9 | ||
| + | |||
| + | Formula: C1340H2150N376O414S9 | ||
| + | Total number of atoms: 4289 | ||
| + | |||
| + | Extinction coefficients: | ||
| + | |||
| + | Extinction coefficients are in units of M-1 cm-1, at 280 nm measured in water. | ||
| + | |||
| + | Ext. coefficient 23045 | ||
| + | Abs 0.1% (=1 g/l) 0.757, assuming all pairs of Cys residues form cystines | ||
| + | |||
| + | |||
| + | Ext. coefficient 22920 | ||
| + | Abs 0.1% (=1 g/l) 0.753, assuming all Cys residues are reduced | ||
| + | |||
| + | Estimated half-life: | ||
| + | |||
| + | The N-terminal of the sequence considered is M (Met). | ||
| + | |||
| + | The estimated half-life is: | ||
| + | 30 hours (mammalian reticulocytes, in vitro). | ||
| + | >20 hours (yeast, in vivo). | ||
| + | >10 hours (Escherichia coli, in vivo). | ||
| + | |||
| + | |||
| + | Instability index: | ||
| + | |||
| + | The instability index (II) is computed to be 34.88 | ||
| + | This classifies the protein as stable. | ||
| + | |||
| + | |||
| + | |||
| + | Aliphatic index: 85.43 | ||
| + | |||
| + | Grand average of hydropathicity (GRAVY): -0.198 | ||
| + | |||
| + | |||
| + | |||
| + | SIB Swiss Institute of Bioinformatics | Disclaimer | ||
| + | |||
| + | ===Source=== | ||
| + | |||
| + | <i>Chlamydomonas reinhardtii</i> | ||
| + | |||
| + | ===References=== | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
| − | <partinfo> | + | <partinfo>BBa_K1080004 parameters</partinfo> |
<!-- --> | <!-- --> | ||
Latest revision as of 01:49, 17 October 2014
ChlM
Mg protoporphyrin IX S-adenosyl methionine O-methyl transferase - Magnesium-protoporphyrin O-methyltransferase (ChlM) [PMID: [http://www.ncbi.nlm.nih.gov/pubmed/12828371 12828371]; [http://www.ncbi.nlm.nih.gov/pubmed/12489983 12489983]; [http://www.ncbi.nlm.nih.gov/pubmed/4436384 4436384]]; ChloroP 1.1 predicts cp location
Operon Usage
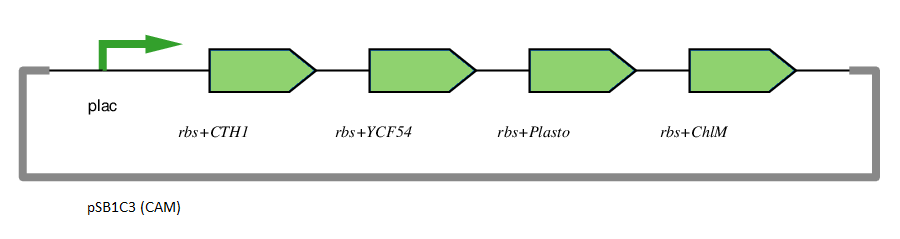
This gene has been used in an operon with other genes responsible for catalysing the biosynthesis pathway from Mg-protoporphyrin IX to Protochlorophyllide. CTH1, Plastocyanin, and YCF54 are involved in the oxidative cyclase pathway. ChlM methylates Mg-protoporphyrin IX, facilitating the highly-regulated catalysis of Mg-chelatase. CTH1 catalyses the conversion of Mg-protoporphyrin IX monomethyl into divinyl protochlorophyllide, interacting with YCF54 and Plastocyanin.
The insertion of magnesium is the key component of the chlorophyll biosynthesis pathway.
The plasmid is under the control of the lac promoter.
Structure
Figure 1: Crystal structure of ChlM from cyanobacteria Synechocystis sp. [http://www.ncbi.nlm.nih.gov/pubmed/25077963]
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 230
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 787
- 1000COMPATIBLE WITH RFC[1000]
Amino Acid Sequence
MASEIAQTAD VGSLTFAVGG VGAVVGLGAL LVATDHQKRR SEQMKSFDGD EKEAVKDYFN
TAGFERWRKI YGETDEVNKV QLDIRTGHAQ TVDKVLRWVD EEGSVQGITV ADCGCGTGSL
AIQLALRGAA VSASDISAAM ASEAEQRYQQ AVAAGQGKAP KVAPKFEALD LESVKGKYDT
VTCLDVMIHY PQDKVDAMIT HLAGLSDRRL IISFAPKTLS YSILKRIGEL FPGPSKATRA
YLHREEDVEA ALKRAGFKVT KREMTATSFY FSRLLEAIRE
References and documentation are available. Please note the modified algorithm for extinction coefficient.
Number of amino acids: 280
Molecular weight: 30440.6
Theoretical pI: 6.25
Amino acid composition:
Ala (A) 36 12.9%
Arg (R) 17 6.1%
Asn (N) 2 0.7%
Asp (D) 18 6.4%
Cys (C) 3 1.1%
Gln (Q) 12 4.3%
Glu (E) 20 7.1%
Gly (G) 23 8.2%
His (H) 5 1.8%
Ile (I) 13 4.6%
Leu (L) 22 7.9%
Lys (K) 19 6.8%
Met (M) 6 2.1%
Phe (F) 10 3.6%
Pro (P) 6 2.1%
Ser (S) 18 6.4%
Thr (T) 17 6.1%
Trp (W) 2 0.7%
Tyr (Y) 8 2.9%
Val (V) 23 8.2%
Pyl (O) 0 0.0%
Sec (U) 0 0.0%
(B) 0 0.0% (Z) 0 0.0% (X) 0 0.0%
Total number of negatively charged residues (Asp + Glu): 38
Total number of positively charged residues (Arg + Lys): 36
Atomic composition:
Carbon C 1340 Hydrogen H 2150 Nitrogen N 376 Oxygen O 414 Sulfur S 9
Formula: C1340H2150N376O414S9 Total number of atoms: 4289
Extinction coefficients:
Extinction coefficients are in units of M-1 cm-1, at 280 nm measured in water.
Ext. coefficient 23045 Abs 0.1% (=1 g/l) 0.757, assuming all pairs of Cys residues form cystines
Ext. coefficient 22920
Abs 0.1% (=1 g/l) 0.753, assuming all Cys residues are reduced
Estimated half-life:
The N-terminal of the sequence considered is M (Met).
The estimated half-life is:
30 hours (mammalian reticulocytes, in vitro).
>20 hours (yeast, in vivo).
>10 hours (Escherichia coli, in vivo).
Instability index:
The instability index (II) is computed to be 34.88 This classifies the protein as stable.
Aliphatic index: 85.43
Grand average of hydropathicity (GRAVY): -0.198
SIB Swiss Institute of Bioinformatics | Disclaimer
Source
Chlamydomonas reinhardtii

