Difference between revisions of "Part:BBa K1321305"
(→Cellulose productivity of G.xylinus ATCC 53582 in HS-glucose) |
|||
| (38 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1321305 short</partinfo> | <partinfo>BBa_K1321305 short</partinfo> | ||
| − | Gluconacetobacter xylinus is an | + | ''Gluconacetobacter xylinus'' is an obligatively aerobic Gram negative bacterium, which produces bacterial cellulose. Strain ATCC 53582 has been reported as one of the highest cellulose-producing strains, and readily produces cellulose with different carbon feedstocks. ''G. xylinus'' can be used as platform for engineering novel protein-cellulose composite biomaterials. |
| + | ''G. xylinus'' ATCC 53582 strain can not be uploaded to the registry due to materials-transfer agreements, however it can be bought from the ATCC culture collection [http://www.lgcstandards-atcc.org/products/all/53582.aspx?geo_country=gb here]. Alternatively, a free closely related Komagataeibacter rhaeticus strain(''K. rhaeticus'' iGEM - part BBa_K1321306) can be accessed, which is a part of the ''G. xylinus'' genetic engineering toolkit (parts BBa_K1321295 - BBa_K1321332). | ||
| − | |||
| − | G.xylinus ATCC 53582 genome | + | '''G. xylinus ATCC 53582 genome sequence''' |
| − | + | G. xylinus ATCC 53582 genome was sequenced as a part of Imperial iGEM 2014 project. The genome is approximately 2.8Mbp in size, with a GC content of 59.82% (see Figure 1). Autoannotation using Prokka (Seemann 2014) identified in total 2491 putative coding sequences, 46 tRNA sequences and 17 rRNA and other RNA sequences. | |
| + | G. xylinus ATCC53582 was sequenced using an Illumina MiSeq platform, with paired-end 250bp reads. After quality control and removal of low-quality reads, a total of approximately 1.02*10^9bp of sequenced data remained, representing approximately 350x coverage of the genome. Usually, a 50x coverage is considered sufficient for successful assembly. | ||
| + | The full genome sequence as annotated contigs are available here [[File:IC14 G.xylinus ATCC53582.zip]]. This file contains the genome sequence as a fasta file and an annotated gff file (Prokka output, which requires Artemis genome browser for viewing). Note, that these files contain contigs, which are not yet ordered into a scaffold. This is a precaution to avoid mis-representation of the large-scale organization of the genome, as the genomes of the closest relatives have proven to be inappropriate for scaffolding. This does not however, affect the quality of the sequences themselves. | ||
| + | [[File:IC14 G.xylinus ATCC 53582 genome overview.jpg|400px|thumb|left|'''Figure 1.''' Overview of the genome of ''G.xylinus'' ATCC 53582. The genome is approximately 2.8Mbp in size, with a GC content of around 60% (internal circle) and approximately 2500 coding sequences (external circle; blue bands denote single CDSs). Note that this overview should be taken as a guide, as further analysis may reveal a more accurate organization of the contigs. ]] | ||
| − | G.xylinus ATCC 53582 growth rate varies depending on the culturing conditions. Growth rate is highest when G.xylinus is cultured at 30degC, 180rpm shaking in a well-aerated setting. However, | + | |
| − | + | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | '''Culturing ''G. xylinus'' ATCC 53582''' | ||
| + | |||
| + | ''G. xylinus'' ATCC 53582 growth rate varies depending on the culturing conditions. Growth rate is highest when ''G. xylinus'' is cultured at 30degC, 180rpm shaking in a well-aerated setting. However, in shaking conditions, the prevalence of cellulose non-producing mutants (cel- mutants) increases, which causes a decrease in the total yield of produced cellulose. For this reason, when high cellulose production is required, ''G. xylinus'' is often grown in static conditions, at 30degC, in a well-aerated setting. In shaking cultures, ATCC 53582 reaches stationary phase in 48-72 hours, whereas in static cultures, depending on the volume of the medium, it can take up to 10 days. When grown on HS-agar plates at 30degC, cellulose-producing colonies become visible within 48-72 hours, cel- mutant colonies between 24 and 48 hours. Cellulose producing colonies can be distinguished from cel- mutants by colony morphology: cel+ have a rough colony morphology, whereas cel- mutants have a smooth colony morphology (see Figure 2). | ||
| + | The most commonly used medium for ''G. xylinus'' is Hestrin-Schramm (HS) medium. HS medium contains the following components (for a protocol for ''G. xylinus'' culturing, see (G. xylinus media and culturing on Imperial 2014 wiki): | ||
2% (w/v) glucose | 2% (w/v) glucose | ||
0.5% (w/v) yeast extract | 0.5% (w/v) yeast extract | ||
| Line 20: | Line 69: | ||
0.15% (w/v) citric acid | 0.15% (w/v) citric acid | ||
| − | [[File:IC14 G.xylinus colony morphology.jpg|400px|thumb|left| | + | [[File:IC14 G.xylinus colony morphology.jpg|400px|thumb|left|'''Figure 2.''' Different colony morphologies of cel+ and cel- ''G. xylinus'' colonies. Cel+ colonies have a rough colony morphology (left), whereas cel- have a smooth colony morphology(right). Plates were inoculated using 50µl of seed cultures grown to stationary phase, and incubated inverted at 30C for 8 days.]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | '''Cellulose productivity of ''G. xylinus'' ATCC 53582 in HS-glucose''' | ||
| + | ATCC 53582 has been reported as one of the highest cellulose-producing strains. ''G. xylinus'' is commonly grown on HS-glucose. However, it can readily use other carbon feedstocks, including sucrose and glycerol (not shown) | ||
| + | (see Figure 3 for pellicle morphology and Figure 4 for comparison of cellulose productivity of ATCC53582 and ''K. rhaeticus'' iGEM strains on HS-glucose medium and experimental details). Because ''G. xylinus'' is an obligate aerobe, cellulose final yield depends strongly on the surface-area-to-volume ratio of the liquid culture it is grown in, as well as aeration efficiency of the system. When grown at 30degC standing, in a 250ml conical flask, sealed with a foam bun, in 50ml HS-glucose medium, ATCC 53582 produces up to 0.6g of cellulose in 5 days (see Figure 4). As 50ml HS-glucose medium contains 1g of glucose, a yield of 0.6g represents 60% glucose to cellulose conversion efficiency. | ||
| + | |||
| + | |||
| + | [[File:IC14 BC pellicle in flasks.jpg|600px|thumb|left|'''Figure 3'''. Bacterial cellulose pellicle made by ATCC 53582. Culture was grown in 30degC standing, using 100ml HS-glucose medium in a 250ml conical flask for 7 days. After 7 days, the culture was autoclaved to kill ''Gluconacetobacter ''cells and stop growth.]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | [[File:IC14 ATCC and iGEM carbon feedstocks experiment update.jpg|600px|thumb|left|'''Figure 4'''. Cellulose productivity of ''K. rhaeticus'' iGEM and ''G. xylinus'' ATCC 53582 strains in HS medium containing different carbon feedstocks. All carbon feedstocks were normalized to 2% (w/v). Cells were cultured in 50ml Corning tubes, filled with 20ml HS medium standing at 30C for 10 days, cultured with loose caps to allow diffusion of air, and kept at 4degC until measurement. Cellulose was washed twice with dH20, treated with 0.1M NaOH for 4 hours at 60degC, washed twice with dH20 and dried at 60degC for 48h before measuring cellulose pellicle weight. N=3, error bars denote SD. ]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | '''Natural antibiotics resistance of ''G. xylinus'' ATCC 53582''' | ||
| + | |||
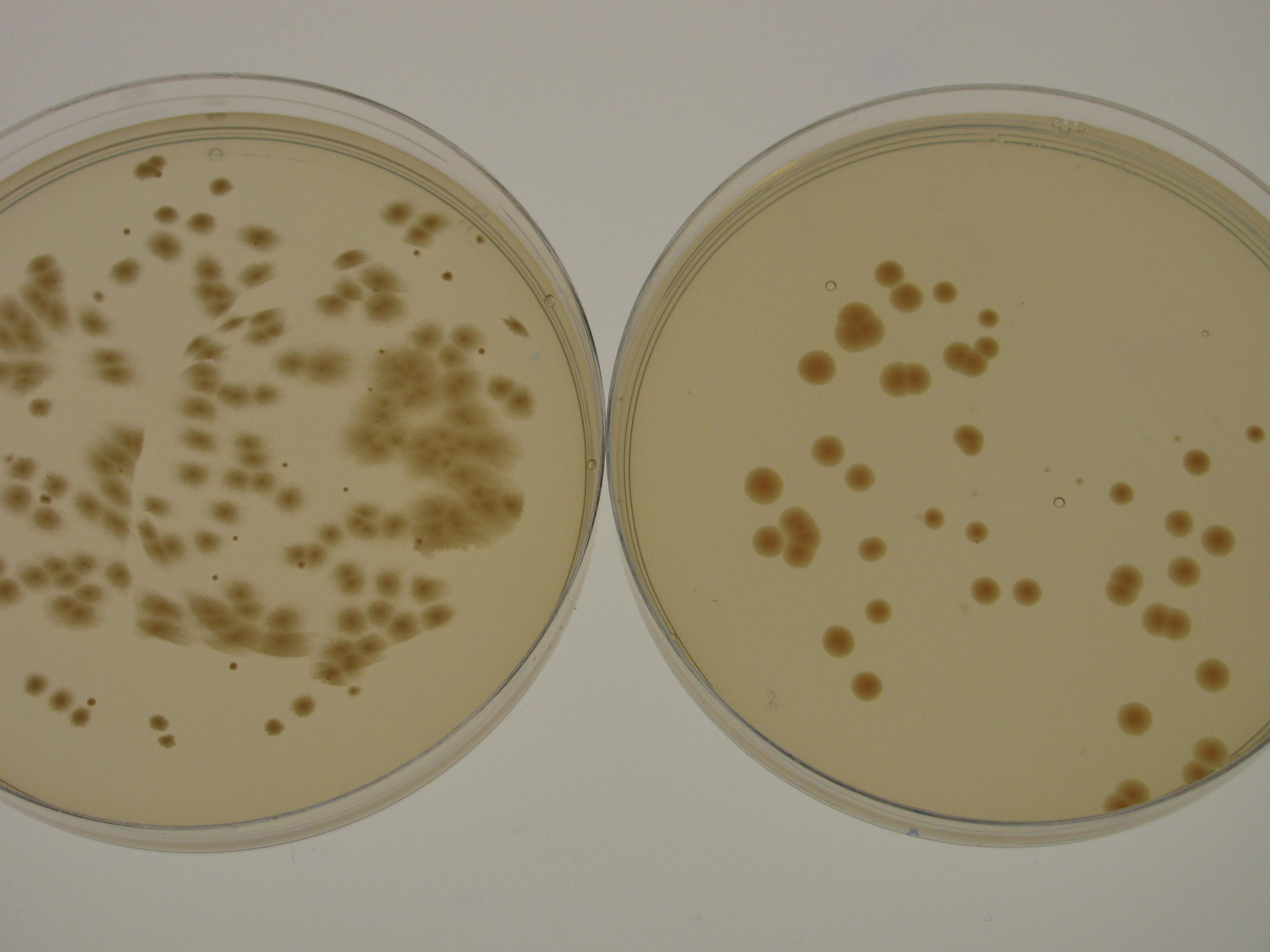
| + | Using optimal antibiotics concentrations is critical for selection of transformed cells while not inhibiting the cell growth. ''G. xylinus'' ATCC 53582 is not resistant to kanamycin and ampicillin in concentrations normally used for ''E. coli'' (50µg/ml and 100µg/ml respectively), however can form colonies on HS-chloramphenicol (at 35µg/ml) when plated at very high concentrations (see Figure 5 for an experiment of ATCC 53582 natural antibiotic concentrations, experimental details, and a comparison of ATCC and iGEM strains). | ||
| + | |||
| + | [[File:IC14 Natural antibiotics resistance of ATCC 53582 and igem strains update.jpg|600px|thumb|left| '''Figure 5.''' Natural antibiotics resistance of ''G. xylinus'' ATCC 53582 and ''K. rhaeticus'' iGEM strains. Due to small colony diameter and large clustering, colonies were rounded to the nearest hundred, if more than a hundred colonies were present. Plates with over 500 colonies often formed a lawn. Plates were seeded with 50µl of ATCC and iGEM seed culture normalized to equal OD600, and grown inverted at 30degC for 5 days until colony counting. N=3, error bars denote SD.]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | '''References:''' | ||
| + | |||
| + | Seemann, T. (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics (Oxford, England). [Online] 30 (14), 2068–2069. Available from: doi:10.1093/bioinformatics/btu153 | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!-- Add more about the biology of this part here | ||
| + | ===Usage and Biology=== | ||
| + | Ref | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 16:47, 6 May 2016
Gluconacetobacter xylinus ATCC 53582
Gluconacetobacter xylinus is an obligatively aerobic Gram negative bacterium, which produces bacterial cellulose. Strain ATCC 53582 has been reported as one of the highest cellulose-producing strains, and readily produces cellulose with different carbon feedstocks. G. xylinus can be used as platform for engineering novel protein-cellulose composite biomaterials.
G. xylinus ATCC 53582 strain can not be uploaded to the registry due to materials-transfer agreements, however it can be bought from the ATCC culture collection [http://www.lgcstandards-atcc.org/products/all/53582.aspx?geo_country=gb here]. Alternatively, a free closely related Komagataeibacter rhaeticus strain(K. rhaeticus iGEM - part BBa_K1321306) can be accessed, which is a part of the G. xylinus genetic engineering toolkit (parts BBa_K1321295 - BBa_K1321332).
G. xylinus ATCC 53582 genome sequence
G. xylinus ATCC 53582 genome was sequenced as a part of Imperial iGEM 2014 project. The genome is approximately 2.8Mbp in size, with a GC content of 59.82% (see Figure 1). Autoannotation using Prokka (Seemann 2014) identified in total 2491 putative coding sequences, 46 tRNA sequences and 17 rRNA and other RNA sequences. G. xylinus ATCC53582 was sequenced using an Illumina MiSeq platform, with paired-end 250bp reads. After quality control and removal of low-quality reads, a total of approximately 1.02*10^9bp of sequenced data remained, representing approximately 350x coverage of the genome. Usually, a 50x coverage is considered sufficient for successful assembly. The full genome sequence as annotated contigs are available here File:IC14 G.xylinus ATCC53582.zip. This file contains the genome sequence as a fasta file and an annotated gff file (Prokka output, which requires Artemis genome browser for viewing). Note, that these files contain contigs, which are not yet ordered into a scaffold. This is a precaution to avoid mis-representation of the large-scale organization of the genome, as the genomes of the closest relatives have proven to be inappropriate for scaffolding. This does not however, affect the quality of the sequences themselves.

Culturing G. xylinus ATCC 53582
G. xylinus ATCC 53582 growth rate varies depending on the culturing conditions. Growth rate is highest when G. xylinus is cultured at 30degC, 180rpm shaking in a well-aerated setting. However, in shaking conditions, the prevalence of cellulose non-producing mutants (cel- mutants) increases, which causes a decrease in the total yield of produced cellulose. For this reason, when high cellulose production is required, G. xylinus is often grown in static conditions, at 30degC, in a well-aerated setting. In shaking cultures, ATCC 53582 reaches stationary phase in 48-72 hours, whereas in static cultures, depending on the volume of the medium, it can take up to 10 days. When grown on HS-agar plates at 30degC, cellulose-producing colonies become visible within 48-72 hours, cel- mutant colonies between 24 and 48 hours. Cellulose producing colonies can be distinguished from cel- mutants by colony morphology: cel+ have a rough colony morphology, whereas cel- mutants have a smooth colony morphology (see Figure 2). The most commonly used medium for G. xylinus is Hestrin-Schramm (HS) medium. HS medium contains the following components (for a protocol for G. xylinus culturing, see (G. xylinus media and culturing on Imperial 2014 wiki): 2% (w/v) glucose 0.5% (w/v) yeast extract 0.5% (w/v) peptone 0.27% (w/v) Na2HPO4 0.15% (w/v) citric acid
Cellulose productivity of G. xylinus ATCC 53582 in HS-glucose ATCC 53582 has been reported as one of the highest cellulose-producing strains. G. xylinus is commonly grown on HS-glucose. However, it can readily use other carbon feedstocks, including sucrose and glycerol (not shown) (see Figure 3 for pellicle morphology and Figure 4 for comparison of cellulose productivity of ATCC53582 and K. rhaeticus iGEM strains on HS-glucose medium and experimental details). Because G. xylinus is an obligate aerobe, cellulose final yield depends strongly on the surface-area-to-volume ratio of the liquid culture it is grown in, as well as aeration efficiency of the system. When grown at 30degC standing, in a 250ml conical flask, sealed with a foam bun, in 50ml HS-glucose medium, ATCC 53582 produces up to 0.6g of cellulose in 5 days (see Figure 4). As 50ml HS-glucose medium contains 1g of glucose, a yield of 0.6g represents 60% glucose to cellulose conversion efficiency.
Natural antibiotics resistance of G. xylinus ATCC 53582
Using optimal antibiotics concentrations is critical for selection of transformed cells while not inhibiting the cell growth. G. xylinus ATCC 53582 is not resistant to kanamycin and ampicillin in concentrations normally used for E. coli (50µg/ml and 100µg/ml respectively), however can form colonies on HS-chloramphenicol (at 35µg/ml) when plated at very high concentrations (see Figure 5 for an experiment of ATCC 53582 natural antibiotic concentrations, experimental details, and a comparison of ATCC and iGEM strains).
References:
Seemann, T. (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics (Oxford, England). [Online] 30 (14), 2068–2069. Available from: doi:10.1093/bioinformatics/btu153

