Difference between revisions of "Part:BBa M0050:Experience"
m (→User Reviews) |
(→Conclusion) |
||
| (16 intermediate revisions by 4 users not shown) | |||
| Line 27: | Line 27: | ||
<I>iGEM14_Edinburgh</I> | <I>iGEM14_Edinburgh</I> | ||
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
| + | |||
| + | ===Caracterisation and contribution iGEM19_Montpellier :=== | ||
| + | |||
| + | <h4><b>Resume :</b></h4> | ||
| + | |||
| + | |||
| + | iGEM19_Montpellier reused this tag based on the work of iGEM14_Edinburgh by adding this ssrA tag to an mRFP1 ([[Part:BBa_M0050]]). A TEV cutting site ([[Part:BBa_J18918]]) has been added between the tag and the mRFP. | ||
| + | The construction of this reporter was inserted under the control of a Bad promoter inducible with arabinose. In order to test the functionality of this reporter system, we first tested the proteolysis activity triggered by the ssrA tag by measuring a decrease in fluorescence between an mRFP alone. After that we looked at whether it was possible to observe a fluorescence gain by separating the ssrA tag. For this we co-expressed the reporter construct (mRFP1-TEVcs-ssrA) and the TEV protease in a NEB10β strain. | ||
| + | |||
| + | |||
| + | <div align="center">[[File:designK3147003.png|650px]]</div> | ||
| + | |||
| + | <div align="center"><b>Figure 1</b>: Construct Design: mRFP1 fused to an ssrA proteolysis tag with a TEV cutting site between the two.</div> | ||
| + | |||
| + | ===Experiment :=== | ||
| + | |||
| + | First we compared the basal fluorescence of the <i>E. coli</i> strain NEB10β transformed with the mRFP1-TEVcs construction and the <i>E. coli</i> NEB10β transformed with the mRFP1-TEVcs-ssrA construction. Our mRFP1-TEVcs is a basal fluorescence control: it has a sequence of the cut TEV cleavage site because in our construction when the TEV separates the ssrA tag, there is still a small end of the cleavage sequence attached to the fluorescent protein. These few amino acids can influence the fluorescence intensity. | ||
| + | |||
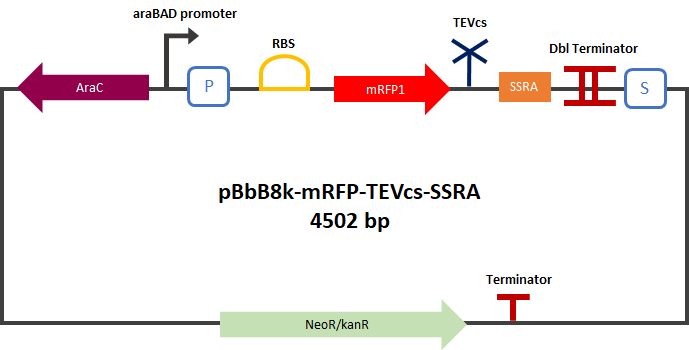
| + | <div align="center">[[File:PlasmideK3147003.png|400px]]</div> | ||
| + | |||
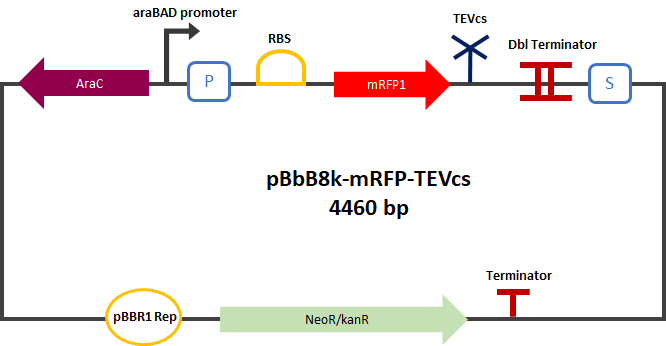
| + | <div align="center">[[File:PlasmideK3147004.png|400px]]</div> | ||
| + | |||
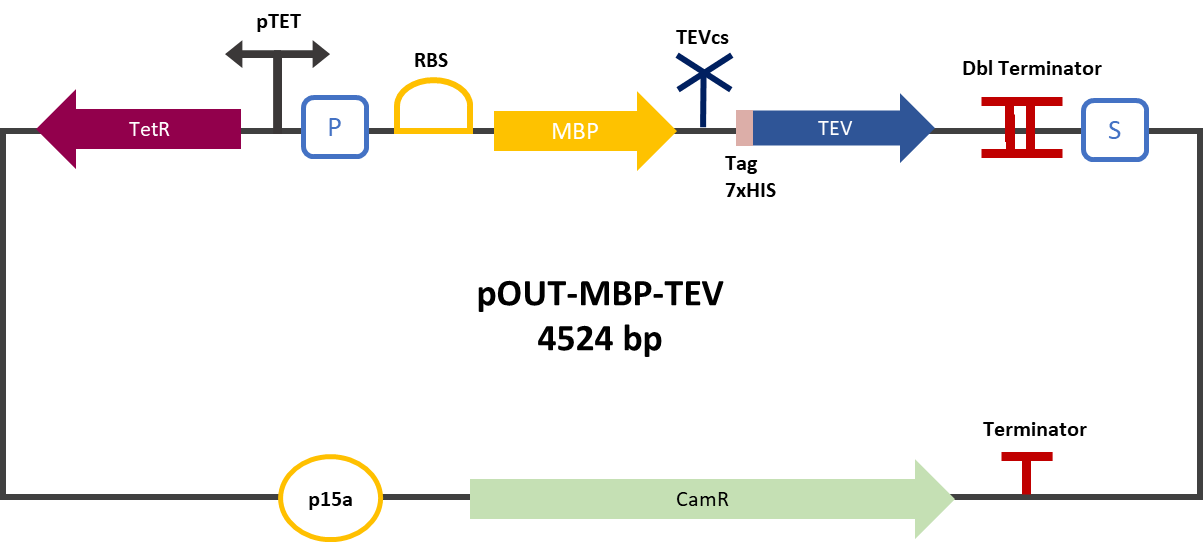
| + | <div align="center">[[File:plasmideK3147005.png|450px]]</div> | ||
| + | |||
| + | <div align="center"><b>Figure 2</b>: mRFP1-TEVcs-ssrA reporter gene in its pBbB8k-GFP backbone; mRFP1-TEVcs reporter gene in its pBbB8k-GFP backbone; MBP-TEV reporter gene in its backbone pOUT18.</div> | ||
| + | |||
| + | The constructions which product the TEV was cloned by Gibson Assembly in a backbone pOUT18 under the control of a TET ON promoter, in order to control its expression. We also added an MBP in N-ter of the protease with a TEV cleavage site in between. The TEV recognition site used for both the protease and the reporter is the classic ENLYFQS site and will be cleaved by the protease when folded [1]. The MBP stabilizes the expression and increases the solubility of the fusion protein [2]. The TEV protease used is optimized for expression in <i>E. coli</i>. The sequence contains a cleavage mutation "S219V anti-self-cleavage". | ||
| + | |||
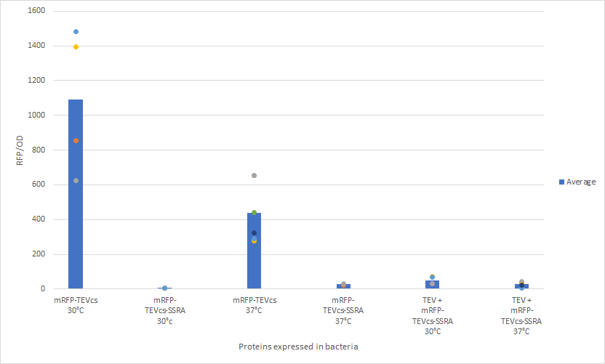
| + | <div align="center">[[File:resultM0050.png|500px]]</div> | ||
| + | |||
| + | <div align="center"><b>Figure 3</b>: Quantification of mRFP-TEVcs constuctions fluorescence with or without a TEV protease</div>. | ||
| + | |||
| + | First, we can see that the ssrA is reducing the mRFP-TEVcs fluorescence strongly (by 99% at 30°C and by 96% at 37°C). Adding a TEV to the proteolysis tag fused construct upgrade the fluorescence by 2 to 4% only, meaning that the TEV action is slower than the ClpX degradation. Also, we can tell from those results that the mRFP-TEVcs is more produced at 30°C. | ||
| + | |||
| + | The constructions that produces the reporter was cloned by Gibson Assembly in a backbone) pBbB8K https://www.addgene.org/35363/ for mRFP1-TEVcs-SSRA under the control of a BAD promoter, in order to control its expression. | ||
| + | In order to carry out this experiment we used <i>E.coli</i> bacteria of the NEB10β strain. Fluorescence data are obtained by a plate reader at 30°C and 37°C. The protease is expressed by inducing the TET promoter with 50ng/mL of aTc (anhydrotetracycline). In this experiment, basal controls of maximum and minimum fluorescence of reporter genes were used. | ||
| + | |||
| + | ===Conclusion=== | ||
| + | |||
| + | The construction mRFP1-TEVcs-ssrA does not seem to be very well expressed from our fluorescence results, however we can see clearly that adding an ssrA tag to our system allows a decrease in the fluorescence of mRFP1. | ||
| + | The low expression of the construction seems to show little significant results concerning the experiment on the separation of the ssrA tag by action of the TEV protease. We therefore carried out a Welch Two sample t-test to verify that the data were significantly different. At 30°C, the p-value we obtained was at 0.0006 while at 37°C it was at 6.5e-07. You can see the graphs of statistical comparison on the main page. | ||
| + | We think that this reporter can be improved by increasing the expression for example, we recommend adding RiboJ [[Part:BBa_K1679038]] and BCD2 [[Part:BBa_J364100]]. The same reporter system was also tested by our team this time by replacing the mRFP1 with an sfGFP, the results are much more relevant, for more information we invite you to visit this page: [[Part:BBa_K3147003]] | ||
| + | For more information on the tests performed with these reporter systems, our results, you can consult our wiki page: https://2019.igem.org/Team:Montpellier/Results. | ||
| + | |||
| + | ==References== | ||
| + | |||
| + | [1] Shih, Y.-P. 2005. « Self-Cleavage of Fusion Protein in Vivo Using TEV Protease to Yield Native Protein ». Protein Science 14(4): 936‑41. | ||
| + | [2] Raran-Kurussi, Sreejith, et David S. Waugh. 2012. « The Ability to Enhance the Solubility of Its Fusion Partners Is an Intrinsic Property of Maltose-Binding Protein but Their Folding Is Either Spontaneous or Chaperone-Mediated » éd. Bostjan Kobe. PLoS ONE 7(11): e49589. | ||
| + | |||
| + | ===iGEM14_Edinburgh=== | ||
| + | |||
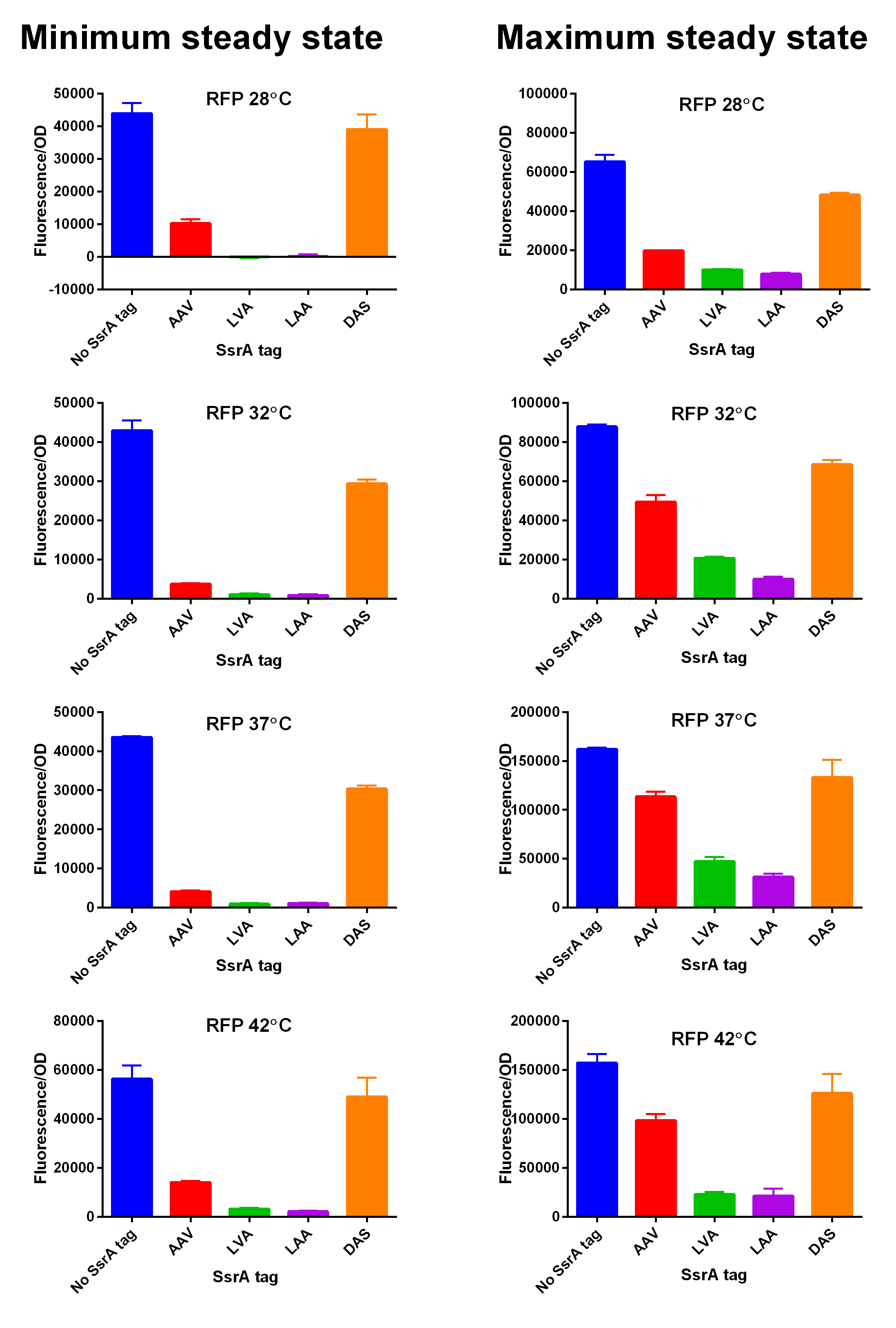
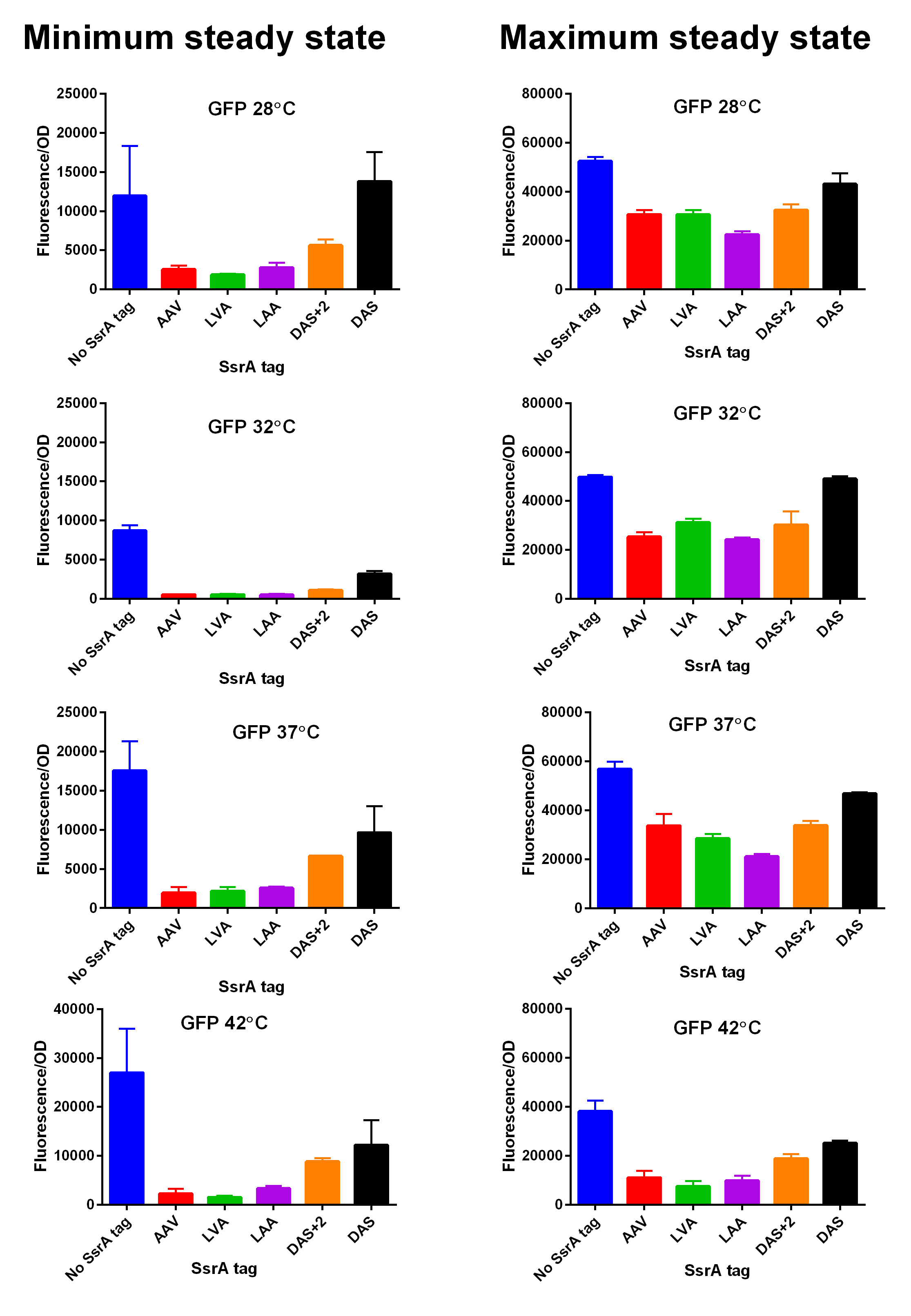
iGEM14_Edinburgh added this tag to RFP ([[Part:BBa_K1399002]]) and GFP ([[Part:BBa_K1399006]]) and constructed lactose/IPTG inducible measurement pathway ([[Part:BBa_K1399012]] and [[Part:BBa_K1399016]], respectively) to characterize and compare the instability of this SsrA-LAA tagged RFP and GFP to others containing different variants of SsrA tags. We determined the steady state fluorescence/OD ratio of E. coli JM109 at uninduced (minimum steady state) and induced (maximum steady state) state. To see how the temperature of environment can affect protein degradation efficiency we repeated experiments in several temperatures (28⁰C, 32⁰C, 37⁰C and 42⁰C). Our data are summarised below. (For more information on SsrA degradation tag system, our experimental assays and results please visit our wiki page [http://2014.igem.org/Team:Edinburgh].) | iGEM14_Edinburgh added this tag to RFP ([[Part:BBa_K1399002]]) and GFP ([[Part:BBa_K1399006]]) and constructed lactose/IPTG inducible measurement pathway ([[Part:BBa_K1399012]] and [[Part:BBa_K1399016]], respectively) to characterize and compare the instability of this SsrA-LAA tagged RFP and GFP to others containing different variants of SsrA tags. We determined the steady state fluorescence/OD ratio of E. coli JM109 at uninduced (minimum steady state) and induced (maximum steady state) state. To see how the temperature of environment can affect protein degradation efficiency we repeated experiments in several temperatures (28⁰C, 32⁰C, 37⁰C and 42⁰C). Our data are summarised below. (For more information on SsrA degradation tag system, our experimental assays and results please visit our wiki page [http://2014.igem.org/Team:Edinburgh].) | ||
[[File:Tagged RFP min and max values at different temperatures.jpg|600px|thumb|center|'''SsrA degron tagged RFP minimum (left) and maximum (right) fluorescence per OD at four different temperatures (28⁰C, 32⁰C, 37⁰C and 42⁰C).''']] [[File:EDiGEM14 RFP legend.png|500px|thumb|center|'''Summary of BioBricks to which our characterization data is applicable.''']] | [[File:Tagged RFP min and max values at different temperatures.jpg|600px|thumb|center|'''SsrA degron tagged RFP minimum (left) and maximum (right) fluorescence per OD at four different temperatures (28⁰C, 32⁰C, 37⁰C and 42⁰C).''']] [[File:EDiGEM14 RFP legend.png|500px|thumb|center|'''Summary of BioBricks to which our characterization data is applicable.''']] | ||
[[File:EdiGEM14 GFP min and max values at different temperatures.jpg|600px|thumb|center|'''SsrA degron tagged GFP minimum (left) and maximum (right) fluorescence per OD at four different temperatures (28⁰C, 32⁰C, 37⁰C and 42⁰C).''']] [[File:EDiGEM14 GFP legend.png|500px|thumb|center|'''Summary of BioBricks to which our characterization data is applicable.''']] | [[File:EdiGEM14 GFP min and max values at different temperatures.jpg|600px|thumb|center|'''SsrA degron tagged GFP minimum (left) and maximum (right) fluorescence per OD at four different temperatures (28⁰C, 32⁰C, 37⁰C and 42⁰C).''']] [[File:EDiGEM14 GFP legend.png|500px|thumb|center|'''Summary of BioBricks to which our characterization data is applicable.''']] | ||
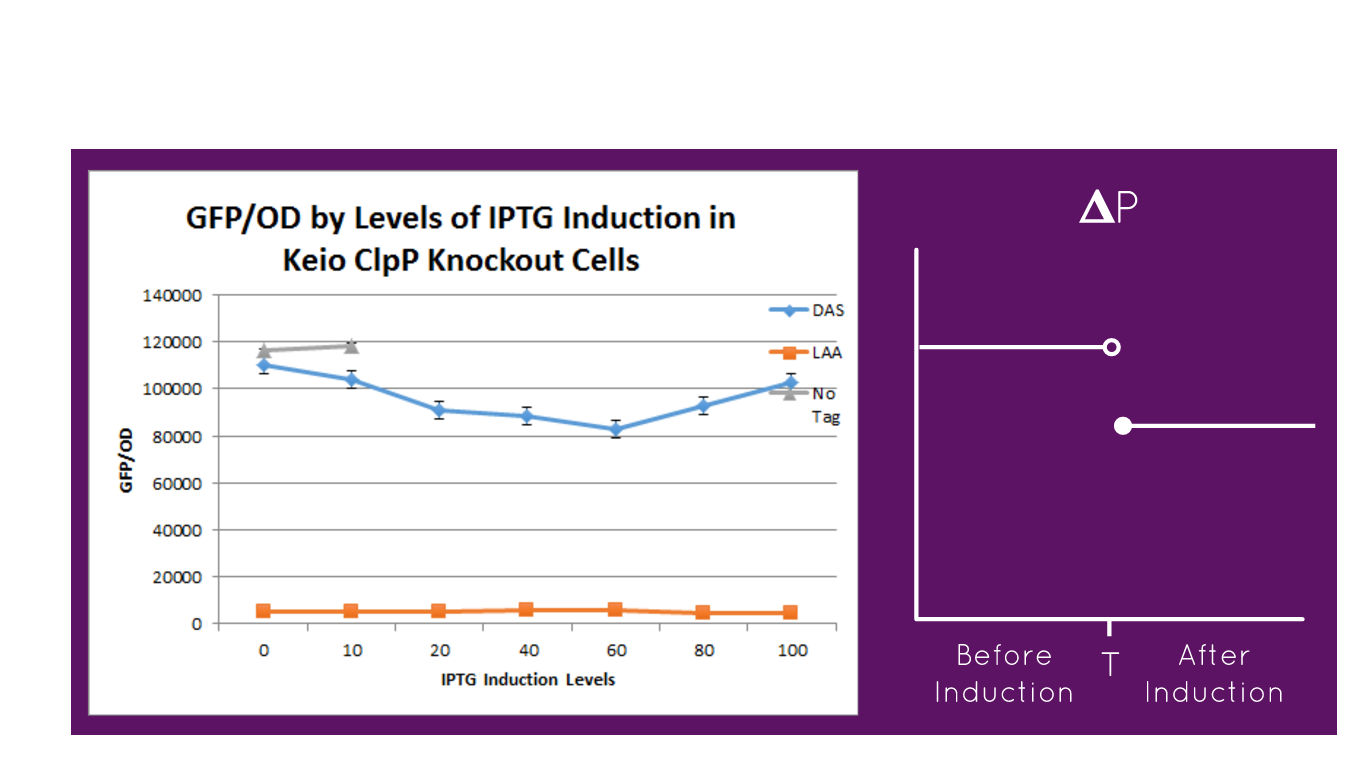
| + | [[File:LAADAS.png|600px]] | ||
| + | <br><br> | ||
| + | The graph depicts the experiment in which our full construct with GFP (no tag, DAS tag, LAA tag) was grown in the Keio ClpP Knockout E. coli strains. Then, it was induced with various IPTG induction levels to allow for the expression of the ClpXP degradation system BBa_K1911001 and the GFP/OD level was observed in each the E. coli cells. The expected results of the experiment were that the no tag would have no decrease in the GFP/OD level as IPTG increased, DAS tag would have a moderate decrease in the GFP/OD level as IPTG increased but not as much of a decrease relative to the Keio Wild Type E. coli cells experiment, and LAA tag would have stronger decrease in the GFP/OD level as IPTG increased but not as much of a decrease relative to the Keio Wild Type E. coli cells experiment. In the actual experiment, the results showed that the GFP/OD level for no tag relatively stayed the same when IPTG increased which was expected, the GFP/OD level for DAS tag decreased when IPTG increased which was expected, and the GFP/OD level for LAA tag decreased more relative to the DAS tag when IPTG increased which was also expected. The no tag data did not have as many data points as the LAA and DAS tag data because there was no growth for the E. coli colonies during the 30-80 micromolar IPTG levels. We theorize this is due to improper inoculation from the colonies. The Keio Wild Type inherently has the ClpXP system existing inside its E. coli cells. The graph above characterizes the BioBrick part M0050 LAA tag because by comparing the degradation of the three different constructs (no tag, DAS tag, LAA tag) we demonstrated that the relative degradation strength of the LAA and DAS tags were indeed what they were supposed to be (LAA- strong, DAS- moderate). | ||
| + | <br><br> | ||
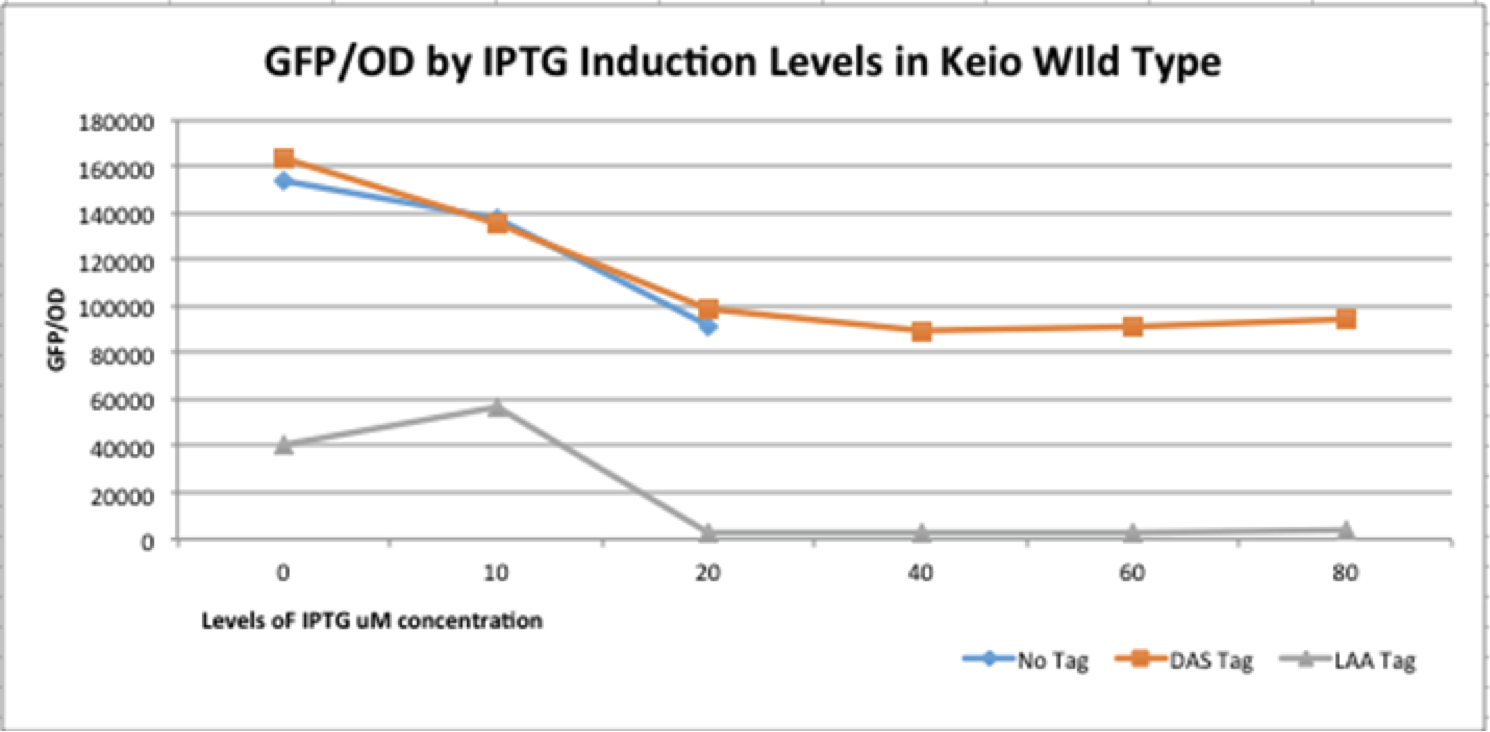
| + | [[File:K365004.png|600px]] | ||
| + | <br><br> | ||
| + | The graph depicts the experiment in which our full construct with GFP (no tag, DAS tag, LAA tag) was grown in the Keio Wild Type E. coli strains. Then, it was induced with various IPTG induction levels to allow for the expression of the ClpXP degradation system BBa_K1911001 and the GFP/OD level was observed in each the E. coli cells. The expected results of the experiment were that the no tag would have no decrease in the GFP/OD level as IPTG increased, DAS tag would have a moderate decrease in the GFP/OD level as IPTG increased, and LAA tag would have stronger decrease in the GFP/OD level as IPTG increased. In the actual experiment, the results showed that the GFP/OD level for no tag decreased when IPTG increased which was not expected, the GFP/OD level for DAS tag decreased when IPTG increased which was expected, and the GFP/OD level for LAA tag decreased more relative to the DAS tag when IPTG increased which was also expected. The no tag data did not have as many data points as the LAA and DAS tag data because there was no growth for the E. coli colonies during the 30-80 micromolar IPTG levels. We theorize this is due to improper inoculation from the colonies. The Keio Wild Type inherently has the ClpXP system existing inside its E. coli cells. The graph above characterizes the BioBrick part M0050 LAA because by comparing the degradation of the three different constructs (no tag, DAS tag, LAA tag) we demonstrated that the relative degradation strength of the LAA and DAS tags were indeed what they were supposed to be (LAA- strong, DAS- moderate) and that the ClpX subunit works in conjunction with the ClpP subunit to degrade the GFP reporter gene. | ||
|} | |} | ||
Latest revision as of 16:57, 20 October 2019
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_M0050
User Reviews
UNIQ70ce4e1a6af04ec7-partinfo-00000000-QINU UNIQ70ce4e1a6af04ec7-partinfo-00000001-QINU
|
••••
iGEM14_Edinburgh |
Caracterisation and contribution iGEM19_Montpellier :Resume :
Figure 1: Construct Design: mRFP1 fused to an ssrA proteolysis tag with a TEV cutting site between the two.
Experiment :First we compared the basal fluorescence of the E. coli strain NEB10β transformed with the mRFP1-TEVcs construction and the E. coli NEB10β transformed with the mRFP1-TEVcs-ssrA construction. Our mRFP1-TEVcs is a basal fluorescence control: it has a sequence of the cut TEV cleavage site because in our construction when the TEV separates the ssrA tag, there is still a small end of the cleavage sequence attached to the fluorescent protein. These few amino acids can influence the fluorescence intensity. Figure 2: mRFP1-TEVcs-ssrA reporter gene in its pBbB8k-GFP backbone; mRFP1-TEVcs reporter gene in its pBbB8k-GFP backbone; MBP-TEV reporter gene in its backbone pOUT18.
The constructions which product the TEV was cloned by Gibson Assembly in a backbone pOUT18 under the control of a TET ON promoter, in order to control its expression. We also added an MBP in N-ter of the protease with a TEV cleavage site in between. The TEV recognition site used for both the protease and the reporter is the classic ENLYFQS site and will be cleaved by the protease when folded [1]. The MBP stabilizes the expression and increases the solubility of the fusion protein [2]. The TEV protease used is optimized for expression in E. coli. The sequence contains a cleavage mutation "S219V anti-self-cleavage". Figure 3: Quantification of mRFP-TEVcs constuctions fluorescence with or without a TEV protease .
First, we can see that the ssrA is reducing the mRFP-TEVcs fluorescence strongly (by 99% at 30°C and by 96% at 37°C). Adding a TEV to the proteolysis tag fused construct upgrade the fluorescence by 2 to 4% only, meaning that the TEV action is slower than the ClpX degradation. Also, we can tell from those results that the mRFP-TEVcs is more produced at 30°C. The constructions that produces the reporter was cloned by Gibson Assembly in a backbone) pBbB8K https://www.addgene.org/35363/ for mRFP1-TEVcs-SSRA under the control of a BAD promoter, in order to control its expression. In order to carry out this experiment we used E.coli bacteria of the NEB10β strain. Fluorescence data are obtained by a plate reader at 30°C and 37°C. The protease is expressed by inducing the TET promoter with 50ng/mL of aTc (anhydrotetracycline). In this experiment, basal controls of maximum and minimum fluorescence of reporter genes were used. ConclusionThe construction mRFP1-TEVcs-ssrA does not seem to be very well expressed from our fluorescence results, however we can see clearly that adding an ssrA tag to our system allows a decrease in the fluorescence of mRFP1. The low expression of the construction seems to show little significant results concerning the experiment on the separation of the ssrA tag by action of the TEV protease. We therefore carried out a Welch Two sample t-test to verify that the data were significantly different. At 30°C, the p-value we obtained was at 0.0006 while at 37°C it was at 6.5e-07. You can see the graphs of statistical comparison on the main page. We think that this reporter can be improved by increasing the expression for example, we recommend adding RiboJ Part:BBa_K1679038 and BCD2 Part:BBa_J364100. The same reporter system was also tested by our team this time by replacing the mRFP1 with an sfGFP, the results are much more relevant, for more information we invite you to visit this page: Part:BBa_K3147003 For more information on the tests performed with these reporter systems, our results, you can consult our wiki page: https://2019.igem.org/Team:Montpellier/Results. References[1] Shih, Y.-P. 2005. « Self-Cleavage of Fusion Protein in Vivo Using TEV Protease to Yield Native Protein ». Protein Science 14(4): 936‑41. [2] Raran-Kurussi, Sreejith, et David S. Waugh. 2012. « The Ability to Enhance the Solubility of Its Fusion Partners Is an Intrinsic Property of Maltose-Binding Protein but Their Folding Is Either Spontaneous or Chaperone-Mediated » éd. Bostjan Kobe. PLoS ONE 7(11): e49589. iGEM14_EdinburghiGEM14_Edinburgh added this tag to RFP (Part:BBa_K1399002) and GFP (Part:BBa_K1399006) and constructed lactose/IPTG inducible measurement pathway (Part:BBa_K1399012 and Part:BBa_K1399016, respectively) to characterize and compare the instability of this SsrA-LAA tagged RFP and GFP to others containing different variants of SsrA tags. We determined the steady state fluorescence/OD ratio of E. coli JM109 at uninduced (minimum steady state) and induced (maximum steady state) state. To see how the temperature of environment can affect protein degradation efficiency we repeated experiments in several temperatures (28⁰C, 32⁰C, 37⁰C and 42⁰C). Our data are summarised below. (For more information on SsrA degradation tag system, our experimental assays and results please visit our wiki page [http://2014.igem.org/Team:Edinburgh].)
|