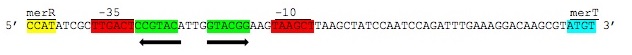Difference between revisions of "Part:BBa K1420004"
| (41 intermediate revisions by 4 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1420004 short</partinfo> | <partinfo>BBa_K1420004 short</partinfo> | ||
| − | |||
| + | <html lang="en"> | ||
| + | <head> | ||
| + | <meta charset="UTF-8"> | ||
| + | <meta name="viewport" content="width=device-width, initial-scale=1.0"> | ||
| + | <title>MerR (BBa_K1420004)</title> | ||
| + | <style> | ||
| + | img { | ||
| + | max-width: 80%; /* Adjust this percentage to change size relative to text */ | ||
| + | height: auto; /* Maintain aspect ratio */ | ||
| + | } | ||
| + | .caption { | ||
| + | text-align: center; | ||
| + | font-size: 0.9em; | ||
| + | margin-top: 5px; | ||
| + | margin-bottom: 20px; /* Space below the caption */ | ||
| + | } | ||
| + | </style> | ||
| + | </head> | ||
| + | <body> | ||
| + | <h2>Improvement made by 2024 Shanghai-city</h2> | ||
| + | <h3>Uploaded by Jiaxi Li<h3> | ||
| + | <h3>Summary</h3> | ||
| + | <p> | ||
| + | We performed codon optimization on MerR and provided a protein structure diagram, named op-merR (BBa_K5068003). At the same time, we will use the optimized op-merR for the detection of mercury ions in the biosensor p15A-op-merR-bspA-pcpS (BBa_K5068007). Our main goal is to improve the sensitivity of biosensors for detecting mercury ions, while also making heavy metal detection methods green and visual to reduce environmental pollution. | ||
| + | </p> | ||
| + | <p> | ||
| + | The <em>bapA</em> and <em>pcpS</em> genes play important roles in the production and transport of pigments. The <em>bapA</em> gene encodes a protein involved in the synthesis of the blue pigment indigo [1], while the <em>pcpS</em> gene encodes a PPTase that activates acyl carrier proteins, facilitating the effective transport of the synthesized pigment within the cell. These two genes typically work in synergy, with <em>bapA</em> synthesizing indigo and <em>pcpS</em> enhancing the transport and stability of the pigment, thereby increasing the bacteria's survival in specific environments [2]. In a heterologous expression system, the combination of <em>bpsA</em> (a single-module non-ribosomal peptide synthetase) and <em>pcpS</em> enables the successful biosynthesis of indigo in <em>Escherichia coli</em>, demonstrating their collaborative role in the pigment biosynthetic pathway. | ||
| + | </p> | ||
| + | <h3>Documentation</h3> | ||
| + | |||
| + | <h4>a. Usage and Biology</h4> | ||
| + | <p> | ||
| + | The op-merR (BBa_K5068003) is a transcriptional regulator responsive to Hg (II) (Fig. 1). The MerR family of metal regulatory factors has been shown to play an important role in heavy metal resistance by regulating the transcription and translation of a series of detoxification-related genes. The extraordinary fidelity, sensing, and regulatory capabilities of the MerR series regulators make them a powerful source of biological components for assembling whole-cell biosensors [3,4]. MerR is a transcription factor belonging to the MerR family, which is used in microorganisms to express detoxification genes and is highly sensitive to specific heavy metal ions in the environment. This transcription factor is targeted at divalent mercury ions and can activate the operon merTPCAD. | ||
| + | </p> | ||
| + | |||
| + | <!-- Figure 1 --> | ||
| + | <div style="text-align:center;"> | ||
| + | <img src="https://static.igem.wiki/teams/5068/bba-k1420004/1.png" alt="Figure 1: Crystal Structure of the Transcription Activator Tn501 MerR in Complex with Hg (II)"> | ||
| + | <div class="caption">Fig. 1. Crystal Structure of the Transcription Activator Tn501 MerR in Complex with Hg (II)</div> | ||
| + | </div> | ||
| + | |||
| + | <p> | ||
| + | The merR-merTPCAD system is responsible for transcriptional activation mediated by Hg (II). In the context of Hg (II) detection biosensors, its role is to initiate the transcription of downstream elements upon the recognition of Hg (II) in the environment (Fig. 2). | ||
| + | </p> | ||
| + | |||
| + | <!-- Figure 2 --> | ||
| + | <div style="text-align:center;"> | ||
| + | <img src="https://static.igem.wiki/teams/5068/bba-k1420004/2.png" alt="Figure 2: Schematic structure of the MerR family transcription activation complex represented by CueR"> | ||
| + | <div class="caption">Fig. 2. Schematic structure of the MerR family transcription activation complex represented by CueR [3-5]</div> | ||
| + | </div> | ||
| + | |||
| + | <h4>b. Characterization</h4> | ||
| + | <p> | ||
| + | We connected the <em>merR</em> and <em>merTPCAD</em> promoters and amplified this fragment by PCR. The length of <em>merR</em> and <em>merTPCAD</em> is 596bp. According to Fig. 3, the target fragment was successfully amplified. | ||
| + | </p> | ||
| + | |||
| + | <!-- Figure 3 --> | ||
| + | <div style="text-align:center;"> | ||
| + | <img src="https://static.igem.wiki/teams/5068/bba-k1420004/3.png" alt="Figure 3: Agarose gel electrophoresis results of DNA fragment amplification"> | ||
| + | <div class="caption">Fig. 3. Agarose gel electrophoresis results of DNA fragment amplification</div> | ||
| + | </div> | ||
| + | |||
| + | <h3>References</h3> | ||
| + | <p> | ||
| + | 1. McNerney MP, Michel CL, Kishore K, Standeven J, Styczynski MP (2019) Dynamic and tunable metabolite control for robust minimal-equipment assessment of serum zinc. <em>Nat Commun</em> 10(1):5514. <a href="https://doi.org/10.1038/s41467-019-13454-1">https://doi.org/10.1038/s41467-019-13454-1</a><br> | ||
| + | 2. Xie Z, Zhang Z, Cao Z, Chen M, Li P, Liu W, Qin H, Zhao X, Tao Y, Chen Y (2017) An external substrate-free blue/white screening system in <em>Escherichia coli</em>. <em>Appl Microbiol Biotechnol</em> 101(9):3811–3820.<br> | ||
| + | 3. Hui C, Guo Y, Zhang W, Gao C, Yang X, Chen Y, Li L, Huang X (2018) Surface display of PbrR on <em>Escherichia coli</em> and evaluation of the bioavailability of lead associated with engineered cells in mice. <em>Sci Rep</em> 8(1):5685. <a href="https://doi.org/10.1038/s41598-018-24134-3">https://doi.org/10.1038/s41598-018-24134-3</a><br> | ||
| + | 4. Hui CY, Guo Y, Yang XQ, Zhang W, Huang XQ (2018) Surface display of metal-binding domain derived from PbrR on <em>E. coli</em> specifically increases lead(II) adsorption. <em>Biotechnol Lett</em> 40(5):837–845. <a href="https://doi.org/10.1007/s10529-018-2533-4">https://doi.org/10.1007/s10529-018-2533-4</a><br> | ||
| + | 5. Kumar S, Verma N, Singh A (2017) Development of cadmium specific recombinant biosensor and its application in milk samples. <em>Sensors Actuators B Chem</em> 240:248–254. | ||
| + | </p> | ||
| + | |||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | ==Summary== | ||
| + | |||
| + | <p>'''•''' ''merR'', located upstream of the rest of the mer operon mercury resistance genes, serves to regulate the ''mer'' operon by activating transcription in the presence of Hg(II) and acting as a weak repressor in the absence of Hg(II).</p> | ||
| + | <p>'''•''' The MerR protein contains a C-terminal effector-binding region that recognizes ionic mercury, forming a homodimer upon binding to Hg(II).</p> | ||
| + | <p>'''•''' The effector binding region of MerR family proteins can vary allowing for great diversity in MerR-like promoters that can respond to a variety of heavy metals as well as antibiotics and oxidative stress.</p> | ||
| + | |||
| + | ==Overview== | ||
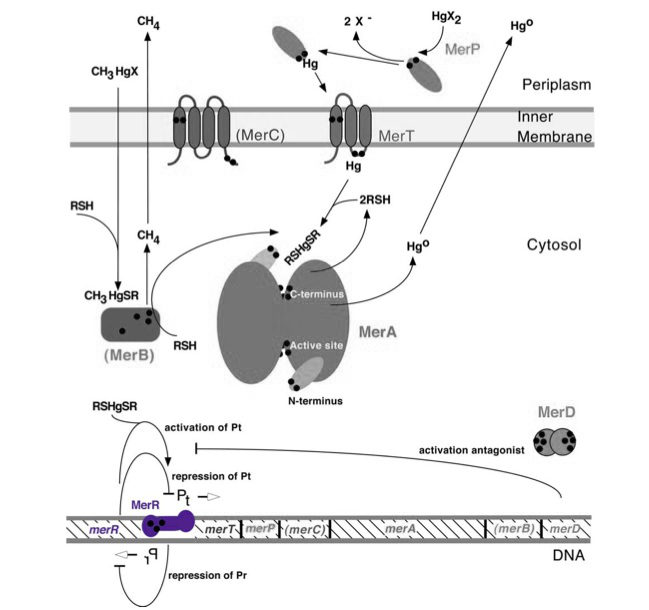
| + | ''merR'', located upstream of the rest of the mer operon mercury resistance genes (see the bottom of ''Figure 1''), serves to regulate the mer operon by activating transcription in the presence of Hg(II) and acting as a weak repressor in the absence of Hg(II). The ''mer'' operon is a set of genes that function in synchrony to convey mercury resistance to bacterial cells. For our purposes the operon can be used to bioremediate methylmercury by converting it to its less toxic ionic form, Hg(II), and then into volatile Hg(0). While variations in the operon exist, with not all genes being present in all organisms and some having extra genes, below is given a general schematic of how the proteins encoded by the operon function to convey mercury resistance to the cell. | ||
| + | |||
| + | |||
| + | [[File:MerR_gene.jpg|center|480px|x]] | ||
| + | |||
| + | ''Figure 1.'' The figure of the mer operon above gives both the key genes and proteins involved in mercury resistance. Black dots in the figure refer to cysteine residues, which may be key in binding mercuric compounds. MerR, highlighted in purple, is the key regulatory protein of the operon, inducing transcription in the presence of ionic mercury. MerT is an inner membrane protein that functions in conjunction with MerP to transport ionic mercury into the cytosol. MerC is a similar inner membrane protein that may not always be necessary for proper functioning of the operon. MerA may be considered the heart of the mercury resistance genes as it converts ionic mercury to the volatile Hg(0) form. For our purposes of bioremediation the function of MerB, which catalyzes the conversion of methylmercury to ionic mercury, is also key. Finally MerD is a putative antagonist of MerR that may also help to regulate the operon. (Figure adapted from "Bacterial mercury resistance from atoms to ecosystems." <sup>1</sup>) | ||
| + | |||
| + | ==Molecular Function and Mechanism== | ||
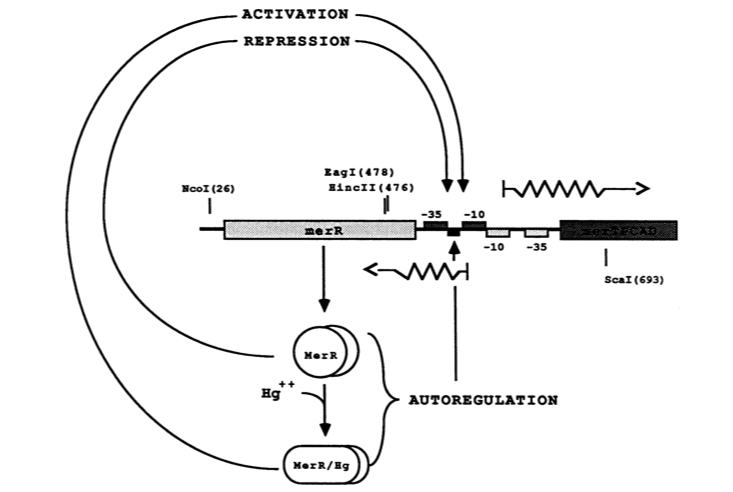
| + | The MerR protein contains a C-terminal effector-binding region that recognizes ionic mercury. Upon binding Hg(II) a MerR homodimer forms. The protein’s N-terminal helix-turn-helix DNA binding domain can now bind the palindromic MerR binding sequence located between the -35 and -10 elements of the PmerT inducible promoter. ''Figure 2'' gives a generalized schematic of the regulatory function of the MerR protein, both for translation of ''merR'' and the entire operon. ''Figure 3'' gives a more detailed outline of the intergenic sequence involved in this conformational change. The promoter has an unusually long spacer sequence of 19 bp that only allows for weak ribosome binding. The MerR/Hg(II) dimer structurally alters the promoter upon binding, allowing for tight DNA polymerase binding and transcription of the downstream genes (''Figure 4''). It should be noted that the effector binding region of MerR family proteins can vary allowing for great diversity in MerR-like promoters that can respond to a variety of heavy metals as well as antibiotics and oxidative stress. This variable nature of MerR family proteins makes them a valuable tool for various heavy metal detection and bioremediation. | ||
| + | |||
| + | |||
| + | |||
| + | [[File:merR_1.TIFF|580px|center|]] | ||
| + | |||
| + | ''Figure 2.'' The figure above shows MerR diamers both in the presence and absence of Hg(II). In both cases the diamer will bind the regulatory intergenic sequence but only when the diamer has undergone a conformational change by binding Hg(II) will transcription be induced, both of merR and the entire operon. (Image adapted from "Genetic analysis of the Tn21 mer operator-promoter." <sup>2</sup>). | ||
| + | |||
| + | |||
| + | <p>[[File:merR_sequence.JPEG ]]</p> | ||
| + | ''Figure 3.'' Intergenic sequences used to regulate the ''mer'' operon. The displayed region is between the ''merR'' and ''merT'' genes containing the -35 and -10 elements of the PmerT inducible promoter highlighted above in red. Between these elements are contained the two palindromic MerR binding sites highlighted in green. Hg(II) bound MerR homodimers bind this region to induce conformational change in the promoter region to allow for DNA polymerase binding and transcription of the ''mer'' operon. (Figure adapted from "Bacterial mercury resistance from atoms to ecosystems." <sup>1</sup>) | ||
| + | <p></p> | ||
| + | |||
| + | |||
| + | <p>[[File:merR_conformation.JPEG ]]</p> | ||
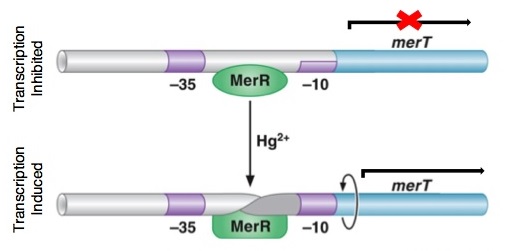
| + | <p>''Figure 4.'' DNA conformational change induced in MerR by Hg(II). This change causes the distortion in the promoter region allowing for DNA polymerase to bind. Without the conformational change, the DNA polymerase bind only weakly to the site. Upon distortion of the promoter region, the polymerase can bind tightly and transcription of the ''mer'' operon is induced. The image also shows how MerR weakly inhibits transcription by binding the site in the absence of mercury, preventing any transcription from occurring. (The image was adapted from "The Mer Operon".<sup>3</sup>)</p> | ||
| + | |||
| + | == References == | ||
| + | <p>1. T. Barkay et al (2003). "Bacterial mercury resistance from atoms to ecosystems." ''FEMS Microbiology Reviews'' <b>27</b>: 355-384.</p> | ||
| + | <p></p> | ||
| + | <p>2. S. J. Park et al (1992). "Genetic analysis of the Tn21 mer operator-promoter." ''J. Bacteriol.'' '''174''': 2160.</p> | ||
| + | <p></p> | ||
| + | <p>3. J. D. Watson (2004). "The Mer Operon." ''Molecular Biology of the Gene.'' New York: Pearson Education. N. pag. Print.</p> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 10: | Line 122: | ||
<!-- --> | <!-- --> | ||
| − | <span class='h3bb'>Sequence and Features</span> | + | <b><span class='h3bb'>Sequence and Features</span></b> |
<partinfo>BBa_K1420004 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1420004 SequenceAndFeatures</partinfo> | ||
Latest revision as of 01:18, 29 September 2024
merR family transcriptional regulator, regulatory protein for mer operon
Improvement made by 2024 Shanghai-city
Uploaded by Jiaxi Li
Summary
Summary
We performed codon optimization on MerR and provided a protein structure diagram, named op-merR (BBa_K5068003). At the same time, we will use the optimized op-merR for the detection of mercury ions in the biosensor p15A-op-merR-bspA-pcpS (BBa_K5068007). Our main goal is to improve the sensitivity of biosensors for detecting mercury ions, while also making heavy metal detection methods green and visual to reduce environmental pollution.
The bapA and pcpS genes play important roles in the production and transport of pigments. The bapA gene encodes a protein involved in the synthesis of the blue pigment indigo [1], while the pcpS gene encodes a PPTase that activates acyl carrier proteins, facilitating the effective transport of the synthesized pigment within the cell. These two genes typically work in synergy, with bapA synthesizing indigo and pcpS enhancing the transport and stability of the pigment, thereby increasing the bacteria's survival in specific environments [2]. In a heterologous expression system, the combination of bpsA (a single-module non-ribosomal peptide synthetase) and pcpS enables the successful biosynthesis of indigo in Escherichia coli, demonstrating their collaborative role in the pigment biosynthetic pathway.
Documentation
a. Usage and Biology
The op-merR (BBa_K5068003) is a transcriptional regulator responsive to Hg (II) (Fig. 1). The MerR family of metal regulatory factors has been shown to play an important role in heavy metal resistance by regulating the transcription and translation of a series of detoxification-related genes. The extraordinary fidelity, sensing, and regulatory capabilities of the MerR series regulators make them a powerful source of biological components for assembling whole-cell biosensors [3,4]. MerR is a transcription factor belonging to the MerR family, which is used in microorganisms to express detoxification genes and is highly sensitive to specific heavy metal ions in the environment. This transcription factor is targeted at divalent mercury ions and can activate the operon merTPCAD.

The merR-merTPCAD system is responsible for transcriptional activation mediated by Hg (II). In the context of Hg (II) detection biosensors, its role is to initiate the transcription of downstream elements upon the recognition of Hg (II) in the environment (Fig. 2).

b. Characterization
We connected the merR and merTPCAD promoters and amplified this fragment by PCR. The length of merR and merTPCAD is 596bp. According to Fig. 3, the target fragment was successfully amplified.

References
1. McNerney MP, Michel CL, Kishore K, Standeven J, Styczynski MP (2019) Dynamic and tunable metabolite control for robust minimal-equipment assessment of serum zinc. Nat Commun 10(1):5514. https://doi.org/10.1038/s41467-019-13454-1
2. Xie Z, Zhang Z, Cao Z, Chen M, Li P, Liu W, Qin H, Zhao X, Tao Y, Chen Y (2017) An external substrate-free blue/white screening system in Escherichia coli. Appl Microbiol Biotechnol 101(9):3811–3820.
3. Hui C, Guo Y, Zhang W, Gao C, Yang X, Chen Y, Li L, Huang X (2018) Surface display of PbrR on Escherichia coli and evaluation of the bioavailability of lead associated with engineered cells in mice. Sci Rep 8(1):5685. https://doi.org/10.1038/s41598-018-24134-3
4. Hui CY, Guo Y, Yang XQ, Zhang W, Huang XQ (2018) Surface display of metal-binding domain derived from PbrR on E. coli specifically increases lead(II) adsorption. Biotechnol Lett 40(5):837–845. https://doi.org/10.1007/s10529-018-2533-4
5. Kumar S, Verma N, Singh A (2017) Development of cadmium specific recombinant biosensor and its application in milk samples. Sensors Actuators B Chem 240:248–254.
Summary
• merR, located upstream of the rest of the mer operon mercury resistance genes, serves to regulate the mer operon by activating transcription in the presence of Hg(II) and acting as a weak repressor in the absence of Hg(II).
• The MerR protein contains a C-terminal effector-binding region that recognizes ionic mercury, forming a homodimer upon binding to Hg(II).
• The effector binding region of MerR family proteins can vary allowing for great diversity in MerR-like promoters that can respond to a variety of heavy metals as well as antibiotics and oxidative stress.
Overview
merR, located upstream of the rest of the mer operon mercury resistance genes (see the bottom of Figure 1), serves to regulate the mer operon by activating transcription in the presence of Hg(II) and acting as a weak repressor in the absence of Hg(II). The mer operon is a set of genes that function in synchrony to convey mercury resistance to bacterial cells. For our purposes the operon can be used to bioremediate methylmercury by converting it to its less toxic ionic form, Hg(II), and then into volatile Hg(0). While variations in the operon exist, with not all genes being present in all organisms and some having extra genes, below is given a general schematic of how the proteins encoded by the operon function to convey mercury resistance to the cell.
Figure 1. The figure of the mer operon above gives both the key genes and proteins involved in mercury resistance. Black dots in the figure refer to cysteine residues, which may be key in binding mercuric compounds. MerR, highlighted in purple, is the key regulatory protein of the operon, inducing transcription in the presence of ionic mercury. MerT is an inner membrane protein that functions in conjunction with MerP to transport ionic mercury into the cytosol. MerC is a similar inner membrane protein that may not always be necessary for proper functioning of the operon. MerA may be considered the heart of the mercury resistance genes as it converts ionic mercury to the volatile Hg(0) form. For our purposes of bioremediation the function of MerB, which catalyzes the conversion of methylmercury to ionic mercury, is also key. Finally MerD is a putative antagonist of MerR that may also help to regulate the operon. (Figure adapted from "Bacterial mercury resistance from atoms to ecosystems." 1)
Molecular Function and Mechanism
The MerR protein contains a C-terminal effector-binding region that recognizes ionic mercury. Upon binding Hg(II) a MerR homodimer forms. The protein’s N-terminal helix-turn-helix DNA binding domain can now bind the palindromic MerR binding sequence located between the -35 and -10 elements of the PmerT inducible promoter. Figure 2 gives a generalized schematic of the regulatory function of the MerR protein, both for translation of merR and the entire operon. Figure 3 gives a more detailed outline of the intergenic sequence involved in this conformational change. The promoter has an unusually long spacer sequence of 19 bp that only allows for weak ribosome binding. The MerR/Hg(II) dimer structurally alters the promoter upon binding, allowing for tight DNA polymerase binding and transcription of the downstream genes (Figure 4). It should be noted that the effector binding region of MerR family proteins can vary allowing for great diversity in MerR-like promoters that can respond to a variety of heavy metals as well as antibiotics and oxidative stress. This variable nature of MerR family proteins makes them a valuable tool for various heavy metal detection and bioremediation.
Figure 2. The figure above shows MerR diamers both in the presence and absence of Hg(II). In both cases the diamer will bind the regulatory intergenic sequence but only when the diamer has undergone a conformational change by binding Hg(II) will transcription be induced, both of merR and the entire operon. (Image adapted from "Genetic analysis of the Tn21 mer operator-promoter." 2).
Figure 3. Intergenic sequences used to regulate the mer operon. The displayed region is between the merR and merT genes containing the -35 and -10 elements of the PmerT inducible promoter highlighted above in red. Between these elements are contained the two palindromic MerR binding sites highlighted in green. Hg(II) bound MerR homodimers bind this region to induce conformational change in the promoter region to allow for DNA polymerase binding and transcription of the mer operon. (Figure adapted from "Bacterial mercury resistance from atoms to ecosystems." 1)
Figure 4. DNA conformational change induced in MerR by Hg(II). This change causes the distortion in the promoter region allowing for DNA polymerase to bind. Without the conformational change, the DNA polymerase bind only weakly to the site. Upon distortion of the promoter region, the polymerase can bind tightly and transcription of the mer operon is induced. The image also shows how MerR weakly inhibits transcription by binding the site in the absence of mercury, preventing any transcription from occurring. (The image was adapted from "The Mer Operon".3)
References
1. T. Barkay et al (2003). "Bacterial mercury resistance from atoms to ecosystems." FEMS Microbiology Reviews 27: 355-384.
2. S. J. Park et al (1992). "Genetic analysis of the Tn21 mer operator-promoter." J. Bacteriol. 174: 2160.
3. J. D. Watson (2004). "The Mer Operon." Molecular Biology of the Gene. New York: Pearson Education. N. pag. Print.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]