Difference between revisions of "Part:BBa K1471001"
| (14 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1471001 short</partinfo> | <partinfo>BBa_K1471001 short</partinfo> | ||
| − | + | Since the project is about phytoremediation, we did codon optimization of merB for its expression in ''Arabidopsis thaliana''. | |
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 17: | Line 17: | ||
<partinfo>BBa_K1471001 parameters</partinfo> | <partinfo>BBa_K1471001 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
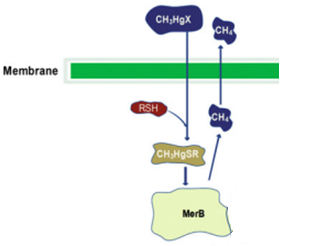
| + | MerB belongs to the mercury resistant bacteria operon, is considered a key enzyme for the bioremediation and detoxification of various mercury compounds; being methylmercury the most notable and relevant. This gene codes for organomercury lyase, catalyzes the protonolysis of the mercury and carbon bond and release less toxic mercury specie (Hg2+). | ||
| + | |||
| + | [[File:Merb.png]] | ||
| + | |||
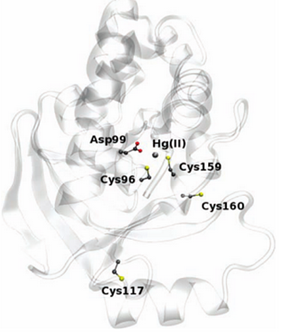
| + | The enzyme has two conserved cysteines residue, Cys-96 and Cys-159, which behave as a substrate binding region. This region has an important role in the cleavage of the carbon-mercury bond. The Asp-99 residue of merB plays an active function in the transference of the proton during the protonolysis. Cys-117 plays the most important structural role. | ||
| + | |||
| + | Other mechanisms have been proposed for the MerB function. One is the mechanism I, this declares that the methylmercury binds first to one of the two cysteines residues, the other cysteine will donate the proton of the leaving group (CH3) in the Hg-C bond cleavage. The mechanism II also refers that the methylmercury binds to one of the cysteines, however the other cysteine transfer the proton to the Asp99. This step allows to the cysteines to coordinate with the methylmercury, then the Asp99 protonates the CH3 and yields the Hg-C cleavage products. | ||
| + | |||
| + | [[File:Merbb.png]] | ||
| + | |||
| + | ===References=== | ||
| + | |||
| + | Comba P. (2011). ''Modeling of Molecular Properties''. Weinheim,Germany:Wiley-VCH. Pages 313-320 | ||
| + | Bizily S., Rugh C., Summers A., Meagher R. (1999). ''Modeling of Molecular Properties''. University of Georgia, USA. | ||
Latest revision as of 14:28, 1 November 2014
MerB.
Since the project is about phytoremediation, we did codon optimization of merB for its expression in Arabidopsis thaliana.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 411
Illegal AgeI site found at 358 - 1000COMPATIBLE WITH RFC[1000]
MerB belongs to the mercury resistant bacteria operon, is considered a key enzyme for the bioremediation and detoxification of various mercury compounds; being methylmercury the most notable and relevant. This gene codes for organomercury lyase, catalyzes the protonolysis of the mercury and carbon bond and release less toxic mercury specie (Hg2+).
The enzyme has two conserved cysteines residue, Cys-96 and Cys-159, which behave as a substrate binding region. This region has an important role in the cleavage of the carbon-mercury bond. The Asp-99 residue of merB plays an active function in the transference of the proton during the protonolysis. Cys-117 plays the most important structural role.
Other mechanisms have been proposed for the MerB function. One is the mechanism I, this declares that the methylmercury binds first to one of the two cysteines residues, the other cysteine will donate the proton of the leaving group (CH3) in the Hg-C bond cleavage. The mechanism II also refers that the methylmercury binds to one of the cysteines, however the other cysteine transfer the proton to the Asp99. This step allows to the cysteines to coordinate with the methylmercury, then the Asp99 protonates the CH3 and yields the Hg-C cleavage products.
References
Comba P. (2011). Modeling of Molecular Properties. Weinheim,Germany:Wiley-VCH. Pages 313-320 Bizily S., Rugh C., Summers A., Meagher R. (1999). Modeling of Molecular Properties. University of Georgia, USA.