Difference between revisions of "Part:BBa K1379007"
Mfcheungaa (Talk | contribs) (→Usage and Biology) |
|||
| (2 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1379007 short</partinfo> | <partinfo>BBa_K1379007 short</partinfo> | ||
| + | <br> This is P<sub>comFA</sub> Measurement Kit [[Part:BBa_K1379003|BBa_K1379003]] assembled with σ<sup>X</sup> generator [[Part:BBa_K1379006|BBa_K1379006]]. | ||
| − | + | ===Usage and Biology=== | |
| − | + | ||
| − | + | P<sub>comFA</sub> (or <i>comFAp</i>) is a σ<sup>X</sup> regulated promoter from <i>Streptococcus pneumoniae</i>. It is a member of the Cin-Box or Com-Box promoter family that share the 8 base pair consensus sequence of TACGAATA, where the sigma factor σ<sup>X</sup> [[Part:BBa_K1379004|BBa_K1379004]] (or ComX) binds to and promotes transcription initiation. (Piotrowski, Luo, & Morrison, 2009) During the exponential growth of <i>S. pneumoniae</i>, Competence Stimulating Peptide (CSP) mediated quorum sensing induces expression of σ<sup>X</sup>. σ<sup>X</sup> as a global regulator then directs <i>S. pneumoniae</i> to enter a transient competent cell state. P<sub>comFA</sub>, alongside with other Com-Box promoters, is turned on by σ<sup>X</sup> and drives expression of the competence protein comFA. (BioCyc) | |
| − | + | Voigt and his colleagues have demonstrated that orthogonal gene expression could be achieved through the use of σs, anti-σs and synthetic promoters. (Rhodius et al., 2013). This principle has been demonstrated to work in <i>E. coli</i> (see characterization below), in which P<sub>comFA</sub> can only be turned on in the presence of σ<sup>X</sup>. | |
| − | < | + | |
| − | + | iGEM 2014 Hong_Kong_HKUST Team has cloned P<sub>comFA</sub> from <i>S. pneumoniae</i> strain NCTC7465 and characterized its Relative Promoter Units (RPU) in presence and absence of a σ<sup>X</sup> generator [[Part:BBa_K1379006|BBa_K1379006]]. Promoter P<sub>celA</sub> ([[Part:BBa_K1379000|BBa_K1379000]]), which is another Com-Box promoter recognized by σ<sup>X</sup>, has also been characterized. | |
| − | + | The transcription start site of this promoter has yet to be experimentally located. | |
| − | + | ||
| − | == Characterization == | + | ===Characterization=== |
| − | + | For characterization, [[Part:BBa_K1379007|BBa_K1379007]] which contain P<sub>comFA</sub> Measurement Kit [[Part:BBa_K1379003|BBa_K1379003]] assembled with σ<sup>X</sup> generator [[Part:BBa_K1379006|BBa_K1379006]] in plasmid [[Part:pSB3K3|pSB3K3]] was used. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
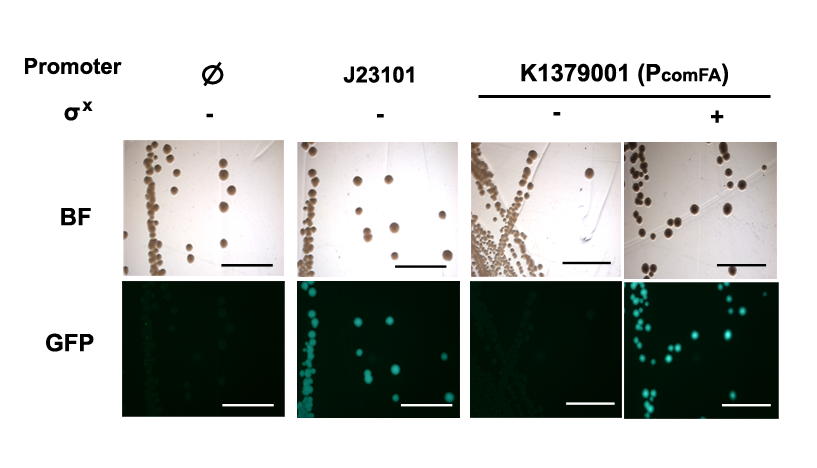
| + | Qualitative characterization was performed by comparing intensities of GFP signals from colonies of <i>E. coli</i> DH10B strain holding the P<sub>comFA</sub> Measurement Kits with and without the σ<sup>X</sup> generator under a fluorescent macroscope with UV filter. Measurement kit for standard reference promoter [[Part:BBa_J23101|BBa_J23101]], [[Part:BBa_I20260|BBa_I20260]] was used as a positive control; [[Part:BBa_E0240|BBa_E0240]] was used as the negative control for background fluorescence. | ||
| + | |||
| + | [[File:pcomFA.png|600px|thumb|center|<b>Figure 1. P<sub>comFA</sub> is on in presence but not absence of σ<sup>X</sup>.</b> P<sub>comFA</sub> Measurement Kit gave strong GFP signals comparable to [[Part:BBa_I20260|BBa_I20260]] when assembled without σ<sup>X</sup> generator, and fluorescence was not observed with the same construct without the σ<sup>X</sup> generator. Scale bar = 5mm.]] | ||
| + | <br> | ||
| + | Quantitative characterization was performed following the protocol described in “Measuring the activity of BioBrick promoters using an <i>in vivo</i> reference standard” (Kelly et al., 2009). <i>E. coli</i> DH10B strains holding the constructs with or without σ<sup>X</sup> generator respectively were grown to mid-log phases. GFP intensities and cell densities were then sampled every 30 minutes for 5 consecutive time points to obtain growth rates and GFP synthesis rates. The GFP synthesis rates were then compared to that of standard reference promoter [[Part:BBa_J23101|BBa_J23101]] measurement device [[Part:BBa_I20260|BBa_I20260]] to obtain the Relative Promoter Units (RPUs). For subtraction of background fluorescence, pSB3K3 holding BBa_E0240 was measured alongside. The measurement was done with 3 replicas. | ||
<br> | <br> | ||
| + | [[File:pcomFAgraph1.png|500px|thumb|center|<b>Figure 2. P<sub>comFA</sub> has 1.21 RPU when paired with σ<sup>X</sup> generator.</b> P<sub>comFA</sub> was measured in reference to [[Part:BBa_J23101|BBa_J23101]] constitutive promoter with and without σ<sup>X</sup> generator [[Part:BBa_K1379006|BBa_K1379006]]. RPU shown was calculated from 3 replicas.]] | ||
| − | + | The above results showed that P<sub>comFA</sub> is functional in <i>E. coli</i> and is turned on specifically in presence of σ<sup>X</sup>. Further characterization can be done to evaluate crosstalk between this sigma factor-promoter pair and their other counterparts. | |
| − | + | ||
| − | < | + | |
| − | + | ||
| − | + | ||
| − | < | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | Extensive documentation of this characterization, including the wet lab protocols and data processing, can be found in [http://2014.igem.org/Team:Hong_Kong_HKUST/pneumosensor/characterization iGEM HKUST 2014 Wiki Page]. | ||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
| Line 59: | Line 37: | ||
<partinfo>BBa_K1379007 parameters</partinfo> | <partinfo>BBa_K1379007 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | |||
| + | ===Reference=== | ||
| + | BioCyc was retrieved from http://biocyc.org/SPNE171101/NEW-IMAGE?type=GENE&object=GJC8-2079 | ||
| + | <br><br> | ||
| + | Luo P., & Morrison D. (2003).'' Transient Association of an Alternative Sigma Factor, ComX, with RNA Polymerase during the Period of Competence for Genetic Transformation in Streptococcus pneumoniae''. Journal of Bacteriology. doi:10.1128/JB.185.1.349-358.2003 | ||
| + | <br><br> | ||
| + | Piotrowski A., Luo P., & Morrison D. (2009). ''Competence for genetic transformation in Streptococcus pneumoniae: termination of activity of the alternative sigma factor ComX is independent of proteolysis of ComX and ComW.'' Journal of Bacteriology. doi:10.1128/JB.01750-08 | ||
| + | <br><br> | ||
| + | Rhodius V., Segall-Shapiro T., Sharon B., Ghodasara A., Orlova E., Tabakh H., . . . Voigt C. (2013). ''Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters.'' Molecular Systhetic Biology .doi:10.1038/msb.2013.58 | ||
| + | <br><br> | ||
| + | J. R. Kelly, A. J. Rubin, J. H. Davis, J. Cumbers, M. J. Czar, ..., D. Endy. (2009). Measuring the activity of BioBrick promoters using an in vivo reference standard. Journal of Biological Engineering, 3, 4. doi: 10.1186/1754-1611-3-4 | ||
Latest revision as of 13:47, 17 October 2014
σx Generator + PcomFA-E0240
This is PcomFA Measurement Kit BBa_K1379003 assembled with σX generator BBa_K1379006.
Usage and Biology
PcomFA (or comFAp) is a σX regulated promoter from Streptococcus pneumoniae. It is a member of the Cin-Box or Com-Box promoter family that share the 8 base pair consensus sequence of TACGAATA, where the sigma factor σX BBa_K1379004 (or ComX) binds to and promotes transcription initiation. (Piotrowski, Luo, & Morrison, 2009) During the exponential growth of S. pneumoniae, Competence Stimulating Peptide (CSP) mediated quorum sensing induces expression of σX. σX as a global regulator then directs S. pneumoniae to enter a transient competent cell state. PcomFA, alongside with other Com-Box promoters, is turned on by σX and drives expression of the competence protein comFA. (BioCyc)
Voigt and his colleagues have demonstrated that orthogonal gene expression could be achieved through the use of σs, anti-σs and synthetic promoters. (Rhodius et al., 2013). This principle has been demonstrated to work in E. coli (see characterization below), in which PcomFA can only be turned on in the presence of σX.
iGEM 2014 Hong_Kong_HKUST Team has cloned PcomFA from S. pneumoniae strain NCTC7465 and characterized its Relative Promoter Units (RPU) in presence and absence of a σX generator BBa_K1379006. Promoter PcelA (BBa_K1379000), which is another Com-Box promoter recognized by σX, has also been characterized.
The transcription start site of this promoter has yet to be experimentally located.
Characterization
For characterization, BBa_K1379007 which contain PcomFA Measurement Kit BBa_K1379003 assembled with σX generator BBa_K1379006 in plasmid pSB3K3 was used.
Qualitative characterization was performed by comparing intensities of GFP signals from colonies of E. coli DH10B strain holding the PcomFA Measurement Kits with and without the σX generator under a fluorescent macroscope with UV filter. Measurement kit for standard reference promoter BBa_J23101, BBa_I20260 was used as a positive control; BBa_E0240 was used as the negative control for background fluorescence.
Quantitative characterization was performed following the protocol described in “Measuring the activity of BioBrick promoters using an in vivo reference standard” (Kelly et al., 2009). E. coli DH10B strains holding the constructs with or without σX generator respectively were grown to mid-log phases. GFP intensities and cell densities were then sampled every 30 minutes for 5 consecutive time points to obtain growth rates and GFP synthesis rates. The GFP synthesis rates were then compared to that of standard reference promoter BBa_J23101 measurement device BBa_I20260 to obtain the Relative Promoter Units (RPUs). For subtraction of background fluorescence, pSB3K3 holding BBa_E0240 was measured alongside. The measurement was done with 3 replicas.
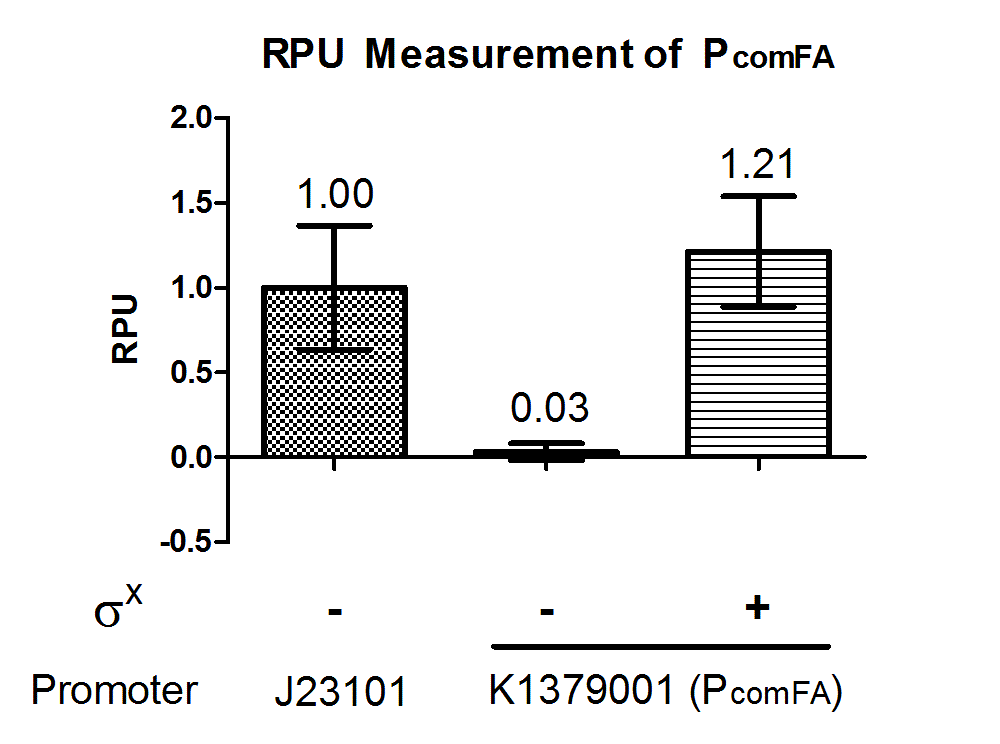
The above results showed that PcomFA is functional in E. coli and is turned on specifically in presence of σX. Further characterization can be done to evaluate crosstalk between this sigma factor-promoter pair and their other counterparts.
Extensive documentation of this characterization, including the wet lab protocols and data processing, can be found in [http://2014.igem.org/Team:Hong_Kong_HKUST/pneumosensor/characterization iGEM HKUST 2014 Wiki Page]. Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 359
Illegal BsaI.rc site found at 1550
Reference
BioCyc was retrieved from http://biocyc.org/SPNE171101/NEW-IMAGE?type=GENE&object=GJC8-2079
Luo P., & Morrison D. (2003). Transient Association of an Alternative Sigma Factor, ComX, with RNA Polymerase during the Period of Competence for Genetic Transformation in Streptococcus pneumoniae. Journal of Bacteriology. doi:10.1128/JB.185.1.349-358.2003
Piotrowski A., Luo P., & Morrison D. (2009). Competence for genetic transformation in Streptococcus pneumoniae: termination of activity of the alternative sigma factor ComX is independent of proteolysis of ComX and ComW. Journal of Bacteriology. doi:10.1128/JB.01750-08
Rhodius V., Segall-Shapiro T., Sharon B., Ghodasara A., Orlova E., Tabakh H., . . . Voigt C. (2013). Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Molecular Systhetic Biology .doi:10.1038/msb.2013.58
J. R. Kelly, A. J. Rubin, J. H. Davis, J. Cumbers, M. J. Czar, ..., D. Endy. (2009). Measuring the activity of BioBrick promoters using an in vivo reference standard. Journal of Biological Engineering, 3, 4. doi: 10.1186/1754-1611-3-4
