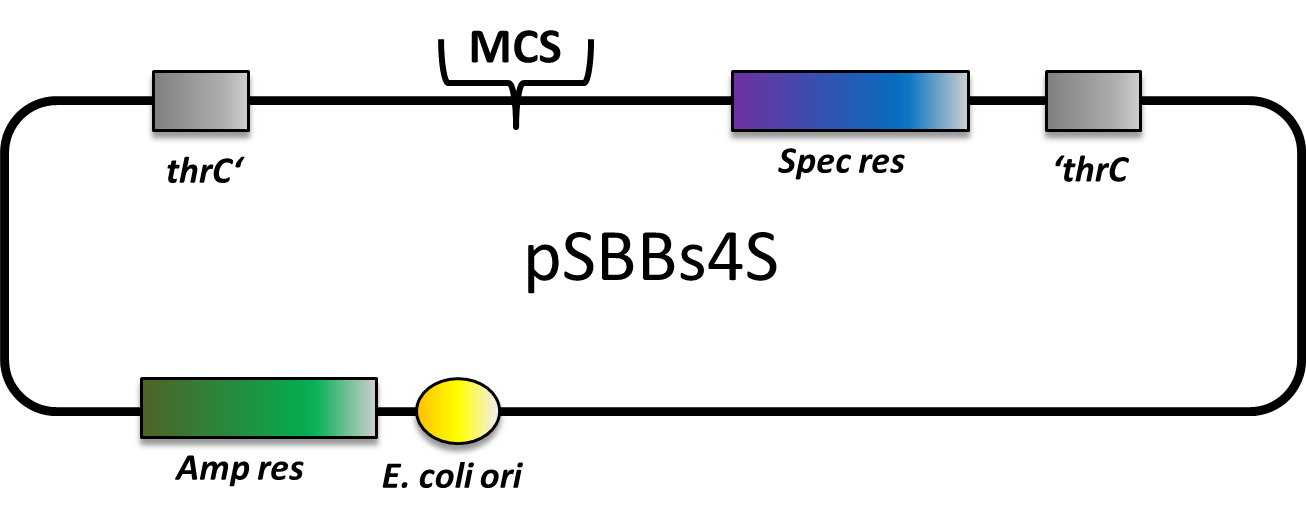Difference between revisions of "Part:BBa J179002"
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_J179002 short</partinfo> | <partinfo>BBa_J179002 short</partinfo> | ||
| − | This part is an empty backbone vector for the usage in Bacillus subtilis. It integrates in the thrC locus and can be selected with Spectinomycin cassette. It has a ampicillin resistance for cloning in E.coli. This backbone is a BioBricked version of the B. subtilis vector pDG1731. | + | This part is an empty backbone vector for the usage in Bacillus subtilis. It integrates in the thrC locus and can be selected with Spectinomycin cassette. It has a ampicillin resistance for cloning in E.coli. This backbone is a ''BioBricked'' version of the ''B. subtilis'' vector pDG1731. |
| + | |||
| + | This version is derived from [https://parts.igem.org/Part:BBa_K823022 BBa_K823022] and contains neither NgoMIV nor AgeI restriction sites, therefore is RFC25-compatible. This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The ''Bacillus'' BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with ''Bacillus subtilis''] by Radeck ''et al.''. | ||
| + | |||
| + | [[Image:LMU-Munich-PSBBs4S.png|600px]] | ||
| + | |||
| + | |||
| + | This BioBrick is part of the LMU 2012 igem project [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks '''''Bacillus''B'''io'''B'''rick'''B'''ox] | ||
| + | |||
| + | For handling of ''B. subtilis'' vectors, please see [https://static.igem.org/mediawiki/2012/c/c4/LMU-Munich_2012_Bacillus_subtilis_vectors.pdf here]. | ||
| + | |||
| + | The transformation into ''B. subtilis'' is explained [https://static.igem.org/mediawiki/2012/4/41/LMU-Munich_2012_Transformation_of_Bacillus_subtilis.pdf here]. | ||
| + | |||
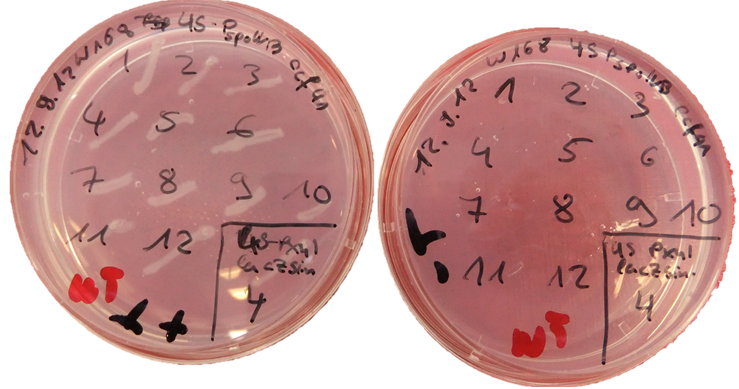
| + | [[Image:LMU-Munich-Thrplate.png| 300px|left]] | ||
| + | We tested the integration of this plasmid with the threonine test: The colonies obtained from the transformation are streaked on three plates: LB with 100mg/L spectinomycine, minimal medium with threonine and minimal medium without threonine. As minimal medium, the MNGE-medium described in the transformation protocol was used. The plates are incubated at 37°C over night. Positive colonies grow on LB and minimal medium with threonine, but not on minimal medium without threonine. | ||
| + | |||
| + | |||
| + | Reference: [http://www.ncbi.nlm.nih.gov/pubmed/8973347 Guérout-Fleury ''et al.''] | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 16:58, 3 February 2014
pBs4S: Bacillus subtilis vector, thrC integration, spec resistance
This part is an empty backbone vector for the usage in Bacillus subtilis. It integrates in the thrC locus and can be selected with Spectinomycin cassette. It has a ampicillin resistance for cloning in E.coli. This backbone is a BioBricked version of the B. subtilis vector pDG1731.
This version is derived from BBa_K823022 and contains neither NgoMIV nor AgeI restriction sites, therefore is RFC25-compatible. This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis] by Radeck et al..
This BioBrick is part of the LMU 2012 igem project [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks BacillusBioBrickBox]
For handling of B. subtilis vectors, please see here.
The transformation into B. subtilis is explained here.
We tested the integration of this plasmid with the threonine test: The colonies obtained from the transformation are streaked on three plates: LB with 100mg/L spectinomycine, minimal medium with threonine and minimal medium without threonine. As minimal medium, the MNGE-medium described in the transformation protocol was used. The plates are incubated at 37°C over night. Positive colonies grow on LB and minimal medium with threonine, but not on minimal medium without threonine.
Reference: [http://www.ncbi.nlm.nih.gov/pubmed/8973347 Guérout-Fleury et al.]
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 4552
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NotI site found at 9
Illegal NotI site found at 4558 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 4552
Illegal BamHI site found at 4534
Illegal XhoI site found at 1011
Illegal XhoI site found at 1929 - 23INCOMPATIBLE WITH RFC[23]Illegal prefix found at 4552
Illegal suffix found at 2 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found at 4552
Plasmid lacks a suffix.
Illegal XbaI site found at 4567
Illegal SpeI site found at 2
Illegal PstI site found at 16 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal BsaI.rc site found at 2856
Illegal SapI.rc site found at 605