Difference between revisions of "Part:BBa K1152007:Experience"
(→General application of BBa_K1152007) |
|||
| (13 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
how you used this part and how it worked out. | how you used this part and how it worked out. | ||
| − | ===General application of BBa_K1152007 | + | <div style="text-align:justify"> |
| + | ===General application of BBa_K1152007=== | ||
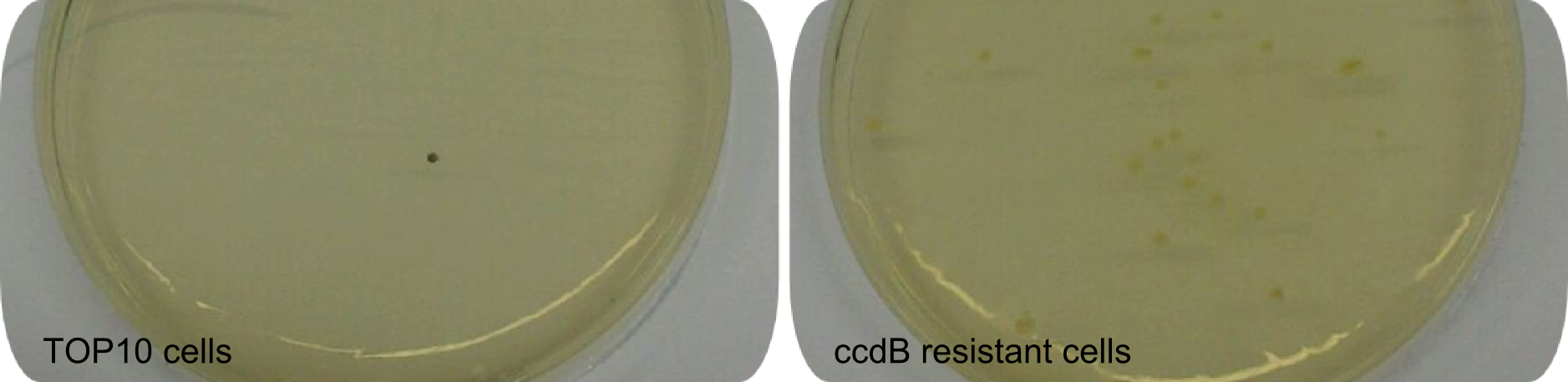
| − | + | BBa_K1152007 was amplified out of four fragments via Gibson Cloning: ccdB, the C-domain of TycC2 and the indC-gene, as well as a backbone. We verified the genotype by colony-PCR, restriction digest with various restriction enzymes, among them EcoRI and PstI, and sequencing of the assembly overhang regions. With the [http://2013.igem.org/Team:Heidelberg/RFCs RFC100] standard, we propose a framework for NRPS-based, customized peptide synthesis. This includes the possibility to tag the desired Non-Ribosomal Peptides with Indigoidine. Using this helper-construct eased our work in bringing the proof of principle for the functionality of the Indigoidine-tag, as by replacing the ccdB with the NRPS modules of interest, re-ligation background is significantly decreased. This construct stands as representative of the various fusion-peptides with Indigoidine-tag that we synthesized during our project. | |
| − | |||
| − | |||
| − | + | [[File:Heidelberg_ccdB_comparison_2.png|500px|center]] | |
| + | <center><div style="width:500px; text-align:justify"><b>Figure 1: Effect of ccdB on non-resistant cells.</b> Regular <em>E. coli</em> TOP10 cells die in presence of active ccdB (left side), while ccdB-resistant cells survive (right side). Hence this comparison shows the effectiveness of ccdB and the minimization of background to about 0%. Using the ccdB-helper plasmid for a Gibson-driven tagging-approach enhances effectiveness significantly. Since cells were not induced with IPTG, colonies did not turn blue.</div></center> | ||
| − | |||
| − | === | + | ===IndC Indigoidine Synthetase ([https://parts.igem.org/Part:BBa_K1152008 BBa_K1152008])=== |
| − | + | An indC expression cassette was transformed into different substrains of E. coli, namely DH5alpha, MG1655, BAP1, TOP10 and NEB Turbo, of which all express the endogenous PPTase entD. This enzyme does not did not show substrate specificity as it was able to activate the T-domain of indC as determined by the blue phenotype of the transformed cells. The transformed cells showed decelerated growth rates upon induction and formed smaller colonies on plates in comparison to negative controls.<br/> | |
| + | Despite decreased growth rates the expression of indC under the endogenous PPTase entD was sufficient for for easy detection of indigoidine production on plates harboring indC-carrying E. coli cells. Therefore, different PPTases were introduced to compare the efficiencies. Co-transformation of host strains with supplementary PPTases led to an improved production of indigoidine. | ||
| − | |||
| − | + | === Application in Peptide-tagging === | |
| − | + | ==== Valine-Indigoidine==== | |
| − | + | ||
| − | ( | + | After amplification of BBa_K1152007 and TycC4dC (which is a module from ''B. parabrevis''), we assembled the Val-Ind-Synthetase using Gibson Cloning. In this approach, ccdB was replaced by the Tyrocidine-module. We verified the genotype by colony-PCR, restriction digest with various restriction enzymes, among them EcoRI and PstI, and sequencing of the assembly overhang regions. Furthermore, SDS-PAGE was performed after induction of cells in order to prove expression – the expected band at 242 kDa was clearly visible after Coomassie staining. Finally, after purification of the NRP, TLC on silica-gel with 100% Dichloromethane as mobile phase, was performed to prove the functionality of the expressed NRPS. |
| − | + | [[Image:Heidelberg_Registry_Fig2.png|800px|center]] | |
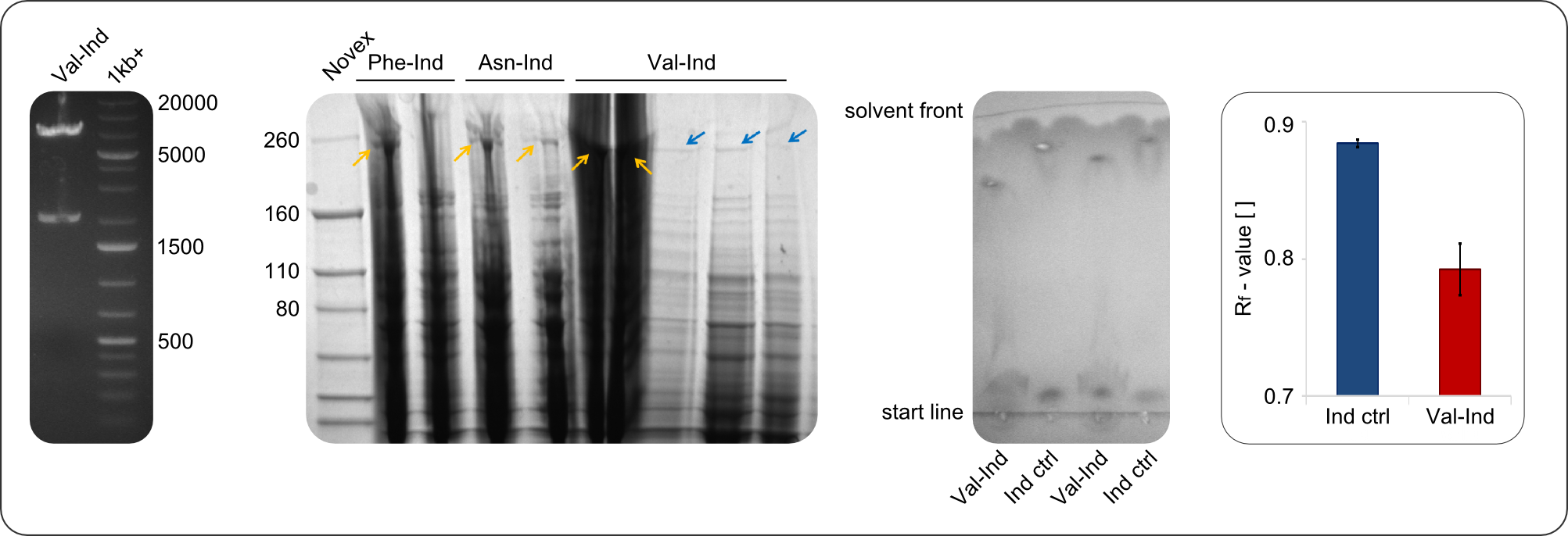
| − | + | <center><div style="width:800px; text-align:justify"><b>Figure 2: Verification of construct.</b> Verification of the genotype (by restriction digest, see first image), the expression (by SDS-PAGE, see second picture) and the functionality (by TLC, see third picture) of <partinfo>BBa_K1152006</partinfo>) and similar constructs. Analyzing the migration behavior of Val-Ind on comparative TLC, a distinct difference to the Indigoidine control is visible.</div></center> | |
| − | |||
| − | |||
| − | + | In restriction digest, as one can see in figure 2, the cutting pattern was as expected. For restriction digest with EcoRI and PstI, as expected two bands were visible, one at around 2000 bp (backbone) and one at around 7000 bp (insert). | |
| − | + | In the SDS-PAGE, expression of the NRPS could be verified, as the expected band at 242kDa is clearly visible after induction with IPTG. To prove that the Val-Ind NRPS is in fact functional, we purified the synthesized NRP and analyzed its properties by comparative TLC with an Indigoidine control. The migration behavior of the synthetic fusion peptide is significantly different from the one of Indigoidine. The samples for Val-Ind tested on TLC were extracted from different clones, hence reproducibility by biological replicates could be proven. | |
| + | ==== Ornithine-Valine-Indigoidine==== | ||
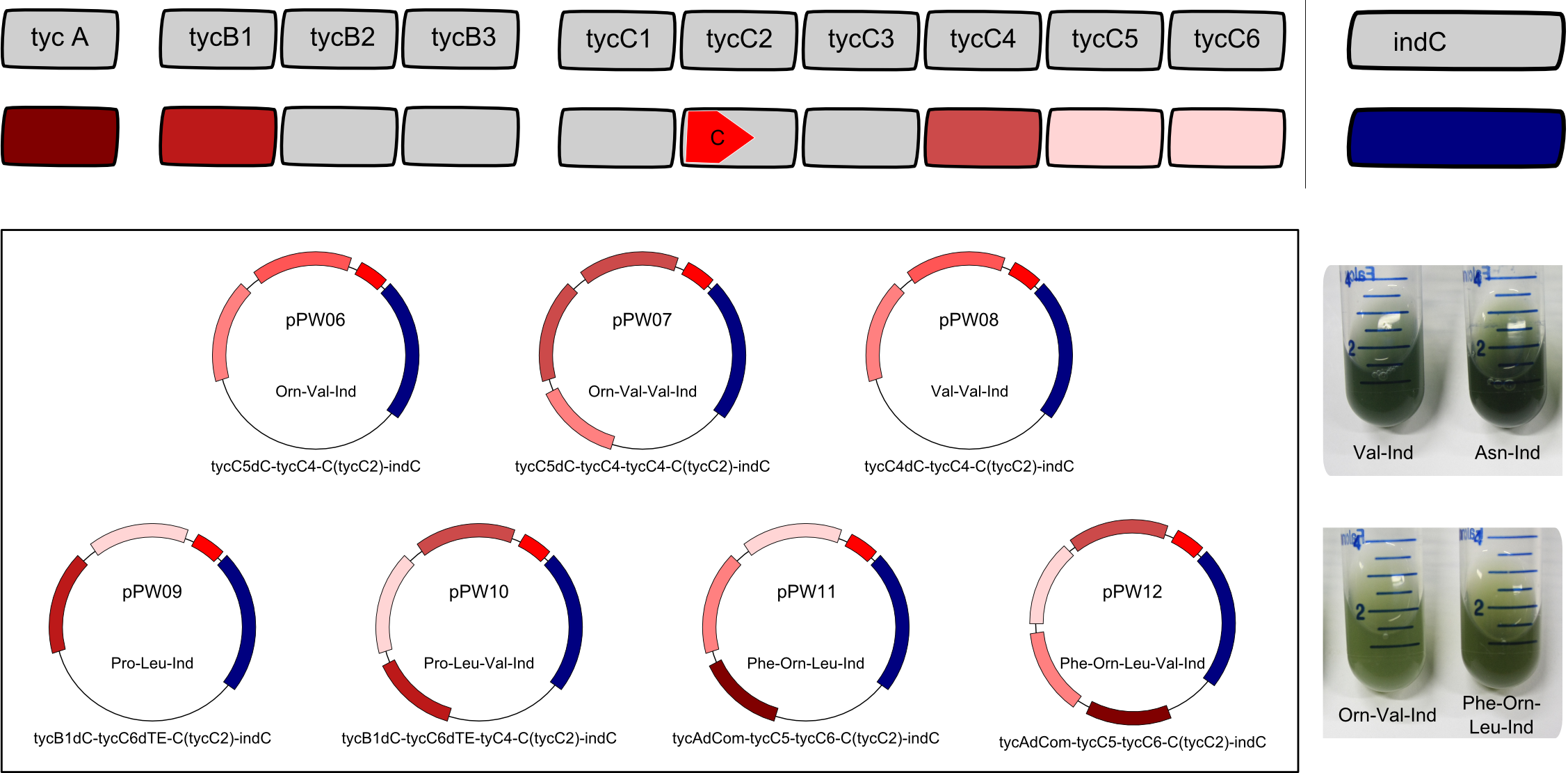
| + | Having shown the functionality of BBa_K1152007, we assembled further constructs with an Indigoidine-tag which can be seen in figure 3. This plasmid (BBa_K1152007) was examined for functionality by TLC only, as this is the very charm of the standard we propose: The '''success''' of an assembly is''' ''visible'' '''to the unaided eye. | ||
| − | |||
| − | [[Image: | + | [[Image:Heidelberg_Registry_2007_Fig.3.png|800px|center]] |
| − | + | ||
| + | <center><div style="width:800px; text-align:justify"><b>Figure 3: Potential of BBa_K1152007.</b> Using the ccdB-helper construct that we provide, tagging of various constructs becomes highly efficient. The tag offers <b>clear visibility</b> - as seen on the image, cultures expressing the NRPS show the characteristic blue phenotype. Furthermore, the tag is <b>inert</b> and <b>universal</b>.</div></center> | ||
| + | |||
| + | |||
| + | |||
| + | By comparative TLC on silica-gel with Dichloromethane as liquid phase, we could show that other synthetases are in fact functional. Furthermore, we could demonstrate that the ccdB-helper-plasmid approach is effective and straightforward. One could imagine easy upscaling of the procedure accompanied by the establishment of a standardized purification protocol that is based on the blue pigment tag. | ||
| + | </div> | ||
===User Reviews=== | ===User Reviews=== | ||
Latest revision as of 18:31, 29 October 2013
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
General application of BBa_K1152007
BBa_K1152007 was amplified out of four fragments via Gibson Cloning: ccdB, the C-domain of TycC2 and the indC-gene, as well as a backbone. We verified the genotype by colony-PCR, restriction digest with various restriction enzymes, among them EcoRI and PstI, and sequencing of the assembly overhang regions. With the [http://2013.igem.org/Team:Heidelberg/RFCs RFC100] standard, we propose a framework for NRPS-based, customized peptide synthesis. This includes the possibility to tag the desired Non-Ribosomal Peptides with Indigoidine. Using this helper-construct eased our work in bringing the proof of principle for the functionality of the Indigoidine-tag, as by replacing the ccdB with the NRPS modules of interest, re-ligation background is significantly decreased. This construct stands as representative of the various fusion-peptides with Indigoidine-tag that we synthesized during our project.
IndC Indigoidine Synthetase (BBa_K1152008)
An indC expression cassette was transformed into different substrains of E. coli, namely DH5alpha, MG1655, BAP1, TOP10 and NEB Turbo, of which all express the endogenous PPTase entD. This enzyme does not did not show substrate specificity as it was able to activate the T-domain of indC as determined by the blue phenotype of the transformed cells. The transformed cells showed decelerated growth rates upon induction and formed smaller colonies on plates in comparison to negative controls.
Despite decreased growth rates the expression of indC under the endogenous PPTase entD was sufficient for for easy detection of indigoidine production on plates harboring indC-carrying E. coli cells. Therefore, different PPTases were introduced to compare the efficiencies. Co-transformation of host strains with supplementary PPTases led to an improved production of indigoidine.
Application in Peptide-tagging
Valine-Indigoidine
After amplification of BBa_K1152007 and TycC4dC (which is a module from B. parabrevis), we assembled the Val-Ind-Synthetase using Gibson Cloning. In this approach, ccdB was replaced by the Tyrocidine-module. We verified the genotype by colony-PCR, restriction digest with various restriction enzymes, among them EcoRI and PstI, and sequencing of the assembly overhang regions. Furthermore, SDS-PAGE was performed after induction of cells in order to prove expression – the expected band at 242 kDa was clearly visible after Coomassie staining. Finally, after purification of the NRP, TLC on silica-gel with 100% Dichloromethane as mobile phase, was performed to prove the functionality of the expressed NRPS.
In restriction digest, as one can see in figure 2, the cutting pattern was as expected. For restriction digest with EcoRI and PstI, as expected two bands were visible, one at around 2000 bp (backbone) and one at around 7000 bp (insert).
In the SDS-PAGE, expression of the NRPS could be verified, as the expected band at 242kDa is clearly visible after induction with IPTG. To prove that the Val-Ind NRPS is in fact functional, we purified the synthesized NRP and analyzed its properties by comparative TLC with an Indigoidine control. The migration behavior of the synthetic fusion peptide is significantly different from the one of Indigoidine. The samples for Val-Ind tested on TLC were extracted from different clones, hence reproducibility by biological replicates could be proven.
Ornithine-Valine-Indigoidine
Having shown the functionality of BBa_K1152007, we assembled further constructs with an Indigoidine-tag which can be seen in figure 3. This plasmid (BBa_K1152007) was examined for functionality by TLC only, as this is the very charm of the standard we propose: The success of an assembly is visible to the unaided eye.
By comparative TLC on silica-gel with Dichloromethane as liquid phase, we could show that other synthetases are in fact functional. Furthermore, we could demonstrate that the ccdB-helper-plasmid approach is effective and straightforward. One could imagine easy upscaling of the procedure accompanied by the establishment of a standardized purification protocol that is based on the blue pigment tag.
User Reviews
UNIQe2bb3d16f826999a-partinfo-00000001-QINU UNIQe2bb3d16f826999a-partinfo-00000002-QINU