Difference between revisions of "Part:BBa K1172904"
(→Barnase) |
(→Usage and Biology) |
||
| (10 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | |||
<br> | <br> | ||
| − | |||
<p align="justify"> | <p align="justify"> | ||
| − | The Barnase (EC 3.1.27) is | + | The Barnase (EC 3.1.27) is a 12 kDa extracellular microbial ribonuclease, which is naturally found in the Gram-positive soil bacteria ''Bacillus amyloliquefaciens'' and consists of a single chain of 110 amino acids. The Barnase (RNase Ba) catalyses the cleavage of single stranded RNA, preferentially behind GpN. In the first step of the RNA-degradation a cyclic intermediate is formed by transesterification and afterwards this intermediate is hydrolyzed yielding in a 3'-nucleotide ([http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_S#References Mossakowska&nscb''et al.'', 1989]).</p> |
<br> | <br> | ||
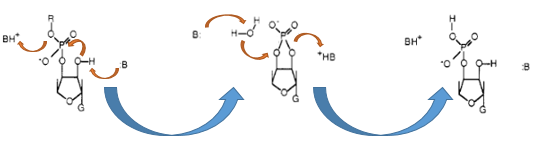
| − | [[Image:IGEM Bielefeld 2013 Biosafety Barnasemove.png|600px|thumb|center|'''Figure 1:''' | + | [[Image:IGEM Bielefeld 2013 Biosafety Barnasemove.png|600px|thumb|center|'''Figure 1:''' Enzymatic reaction of the RNA-cleavage by the RNase Ba. First the transesterification by the Glu-73 residue is performed and then this cyclic intermediate is hydrolyzed by the His-102 of the Barnase]] |
<br> | <br> | ||
<p align="justify"> | <p align="justify"> | ||
| − | In ''Bacillus amyloliquefaciens'' the activity | + | In ''Bacillus amyloliquefaciens'', the activity of the Barnase (RNase Ba) is inhibited intracellular by an inhibitor called barstar. Barstar consists of only 89 amino acids and binds with a high affinity to the toxic Barnase. This prevents the cleavage of the intracellular RNA in the host organism ([http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_S#References Paddon ''et al.'', 1989]). Therefore the Barnase normally acts only outside the cell and is translocated under natural conditions. For this BioBrick we modified the enzyme by cloning only the sequence responsible for the cleavage of the RNA, leaving out the N-terminal signal peptide part.</p> |
<br> | <br> | ||
| − | <span class='h3bb'>Sequence and Features</span> | + | <br> |
| + | <br> | ||
| + | <br> | ||
| + | <span class='h3bb'>'''Sequence and Features'''</span> | ||
<partinfo>BBa_K1172904 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1172904 SequenceAndFeatures</partinfo> | ||
<br> | <br> | ||
| − | As shown in | + | <p align="justify"> |
| − | + | As shown in Figure 2 below, the transcription of the DNA, which encodes the Barnase produces a 474 nt RNA. The translation of the RNA starts about 25 nucleotides downstream from the transcription start and can be divided into two parts. The first part (colored in orange) is translated into a signal peptide at the amino-terminus of the Barnase coding RNA. This part is responsible for the extracellular translocation of the RNase Ba, while the peptide sequence for the active Barnase starts 142 nucleotides downstream from the transcription start (colored in red).<br> | |
| + | The BioBrick (<bbpart>BBa_K1172904</bbpart>) contains only the part of the Barnase encoding the catalytic domain without the extracellular translocation signal of the toxic gene product. Translation of the barnase gene leads to rapid cell death if the expression of the Barnase is not repressed.</p> | ||
<br> | <br> | ||
| − | [[Image: | + | [[Image:Team-Bielefeld-Biosafety_Barnase_Sequence.png|500px|thumb|center|'''Figure 2:''' Sequence of the signal peptide amino terminal of the RNase Ba (Barnase). The Biobrick <bbpart>BBa_K1172904</bbpart> does not contain the signal sequence for the extracellular translocation, but only the coding sequence for the mature enzyme.]] |
| − | + | ||
| + | ===References=== | ||
| + | *Mossakowska, Danuta E. Nyberg, Kerstin and Fersht, Alan R. (1989) Kinetic Characterization of the Recombinant Ribonuclease from Bacillus amyloliquefaciens (Barnase) and Investigation of Key Residues in Catalysis by Site-Directed Mutagenesis [http://pubs.acs.org/doi/pdf/10.1021/bi00435a033|''Biochemistry 28: 3843 - 3850.'']. | ||
| + | *Paddon, C. J. Vasantha, N. and Hartley, R. W. (1989) Translation and Processing of Bacillus amyloliquefaciens Extracellular Rnase [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC209718/pdf/jbacter00168-0575.pdf|''Journal of Bacteriology 171: 1185 - 1187.'']. | ||
| + | *Voss, Carsten Lindau, Dennis and Flaschel, Erwin (2006) Production of Recombinant RNase Ba and Its Application in Downstream Processing of Plasmid DNA for Pharmaceutical Use [http://onlinelibrary.wiley.com/doi/10.1021/bp050417e/pdf|''Biotechnology Progress 22: 737 - 744.'']. | ||
<br><br> | <br><br> | ||
Latest revision as of 14:03, 30 October 2013
Rnase Ba (Barnase) from Bacillus amyloliquefaciens
Usage and Biology
The Barnase (EC 3.1.27) is a 12 kDa extracellular microbial ribonuclease, which is naturally found in the Gram-positive soil bacteria Bacillus amyloliquefaciens and consists of a single chain of 110 amino acids. The Barnase (RNase Ba) catalyses the cleavage of single stranded RNA, preferentially behind GpN. In the first step of the RNA-degradation a cyclic intermediate is formed by transesterification and afterwards this intermediate is hydrolyzed yielding in a 3'-nucleotide ([http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_S#References Mossakowska&nscbet al., 1989]).
In Bacillus amyloliquefaciens, the activity of the Barnase (RNase Ba) is inhibited intracellular by an inhibitor called barstar. Barstar consists of only 89 amino acids and binds with a high affinity to the toxic Barnase. This prevents the cleavage of the intracellular RNA in the host organism ([http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_S#References Paddon et al., 1989]). Therefore the Barnase normally acts only outside the cell and is translocated under natural conditions. For this BioBrick we modified the enzyme by cloning only the sequence responsible for the cleavage of the RNA, leaving out the N-terminal signal peptide part.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
As shown in Figure 2 below, the transcription of the DNA, which encodes the Barnase produces a 474 nt RNA. The translation of the RNA starts about 25 nucleotides downstream from the transcription start and can be divided into two parts. The first part (colored in orange) is translated into a signal peptide at the amino-terminus of the Barnase coding RNA. This part is responsible for the extracellular translocation of the RNase Ba, while the peptide sequence for the active Barnase starts 142 nucleotides downstream from the transcription start (colored in red).
The BioBrick (BBa_K1172904) contains only the part of the Barnase encoding the catalytic domain without the extracellular translocation signal of the toxic gene product. Translation of the barnase gene leads to rapid cell death if the expression of the Barnase is not repressed.

References
- Mossakowska, Danuta E. Nyberg, Kerstin and Fersht, Alan R. (1989) Kinetic Characterization of the Recombinant Ribonuclease from Bacillus amyloliquefaciens (Barnase) and Investigation of Key Residues in Catalysis by Site-Directed Mutagenesis [http://pubs.acs.org/doi/pdf/10.1021/bi00435a033|Biochemistry 28: 3843 - 3850.].
- Paddon, C. J. Vasantha, N. and Hartley, R. W. (1989) Translation and Processing of Bacillus amyloliquefaciens Extracellular Rnase [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC209718/pdf/jbacter00168-0575.pdf|Journal of Bacteriology 171: 1185 - 1187.].
- Voss, Carsten Lindau, Dennis and Flaschel, Erwin (2006) Production of Recombinant RNase Ba and Its Application in Downstream Processing of Plasmid DNA for Pharmaceutical Use [http://onlinelibrary.wiley.com/doi/10.1021/bp050417e/pdf|Biotechnology Progress 22: 737 - 744.].