Difference between revisions of "Part:BBa K1211001"
(→Data) |
Alisaleong (Talk | contribs) |
||
| (19 intermediate revisions by 4 users not shown) | |||
| Line 3: | Line 3: | ||
This is the DNA coding for the enzyme propionate CoA transferase from Clostridum propionicum. The enzyme normally catalyzes the reaction: acetyl-CoA + propanoate <--> acetate + propanoyl-CoA. This mutated sequence has four point mutations and one amino acid substitution (compared to the wild type enzyme) in order to produce (D)-Lactyl-CoA. <br> | This is the DNA coding for the enzyme propionate CoA transferase from Clostridum propionicum. The enzyme normally catalyzes the reaction: acetyl-CoA + propanoate <--> acetate + propanoyl-CoA. This mutated sequence has four point mutations and one amino acid substitution (compared to the wild type enzyme) in order to produce (D)-Lactyl-CoA. <br> | ||
| − | We used this enzyme as a the first enzyme in the pathway to create PLA or Polylactic acid. However, this biobrick | + | <br> |
| − | + | '''We used this enzyme as a the first enzyme in the pathway to create PLA or Polylactic acid. However, this biobrick produces (D)-Lactyl-CoA, which is the precursor to all PHAs, and thus this biobrick can be used in a wide range of projects in the future.''' | |
| Line 14: | Line 14: | ||
| − | *Our project used this gene along with a ''Pseudomonas resinovorans'' PHA synthase gene to produce PLA. We tested the ability of our ''E. coli'' to produce PLA by using Nile red | + | *Our project used this gene along with a ''Pseudomonas resinovorans'' PHA synthase gene to produce PLA. However, other teams can add other heterologous enzymes and produce different PHA. We tested the ability of our ''E. coli'' to produce PLA by using Nile red |
**Nile red is an intercellular lipid strain | **Nile red is an intercellular lipid strain | ||
**Nile red does not affect the growth of bacteria, and its fluorescence is quenched in water | **Nile red does not affect the growth of bacteria, and its fluorescence is quenched in water | ||
| − | *We then proceeded to test our plasmid in the plate reader. | + | *We then proceeded to test our strain with out plasmid in the plate reader. |
| − | **Cells were grown for 24 hours with both enzymes induced and in the presence of Nile red. The cells were washed and re-suspended in PBS. The readings were normalized for optical density. | + | **Cells were grown for 24 hours with both enzymes induced and in the presence of Nile red. The cells were washed and re-suspended in PBS. The readings were normalized for optical density. Clearly, the one with our plasmid is more fluorescent indicating PLA production. |
<center>[[File:EcNR2 LB x20 wiki.jpg|400px]]</center><br><br> | <center>[[File:EcNR2 LB x20 wiki.jpg|400px]]</center><br><br> | ||
| + | |||
| + | *After creating diversity using MAGE or Multiplex automated genome engineering, we needed a way to sort out those with the highest levels of fluorescence indicating greater PLA production. We decided to use FACS or FLuorescence activated cell sorting, to select those cells with the highest levels of fluorescence. | ||
{| | {| | ||
| − | |The first sample is the wild type cells. These do not have any heterologous enzymes and thus should not show any significant Nile red fluorescence, since they are not producing PLA | + | |The first sample is the wild type cells. These do not have any heterologous enzymes and thus should not show any significant Nile red fluorescence, since they are not producing PLA. To keep all the control in place, this strain was grown overnight with the inducers and in the presence of Nile red to strain the PLA (this is the same procedure for all later strains as well). |
|style="padding-left: 20px; padding-right: 20px;"|[[File:wildtypea.jpg|400px]] | |style="padding-left: 20px; padding-right: 20px;"|[[File:wildtypea.jpg|400px]] | ||
|} | |} | ||
| Line 30: | Line 32: | ||
{| | {| | ||
| − | |This is EcNR2 with our plasmid containing both the PCT and PHA gene | + | |This is EcNR2 with our plasmid containing both the PCT and PHA gene. The gate (labeled P2) was chosen to select those with the highest levels of fluorescence. There is clearly a bimodal distribution that appeared when our construct was added. We predict that this smaller peak represents those cells who were able to produce enough PLA to be detected by FACS. |
|style="padding-left: 20px; padding-right: 20px;"|[[File:baselinereal.jpg|400px]] | |style="padding-left: 20px; padding-right: 20px;"|[[File:baselinereal.jpg|400px]] | ||
|} | |} | ||
| Line 37: | Line 39: | ||
{| | {| | ||
|This is a sample with sample is the all oligos. This is a combination of both the RBS oligos and the KO oligos. This sample has 98 cells within the gate, which is a six and a half fold increase. This means that there was diversity in our population and thus MAGE worked to increase the amount of PLA that our ''E. coli'' could produce. | |This is a sample with sample is the all oligos. This is a combination of both the RBS oligos and the KO oligos. This sample has 98 cells within the gate, which is a six and a half fold increase. This means that there was diversity in our population and thus MAGE worked to increase the amount of PLA that our ''E. coli'' could produce. | ||
| − | |style="padding-left: 20px; padding-right: 20px;"|[[File:66-All both.jpg]] | + | |style="padding-left: 20px; padding-right: 20px;"|[[File:66-All both.jpg|400px]] |
|} | |} | ||
| + | To read more about our project [http://2013.igem.org/Team:Yale/Project_Overview click here] <br> | ||
| + | [[File:PLA pathway2.jpg|600px]] | ||
| + | <br> | ||
| + | <html> | ||
| + | <h2>Team PuiChing Macau 2021: Optimization and Plastic Production</h2> | ||
| + | Based on the previous construct of Yale 2013 (BBa_K1211001), we have optimized the sequence(the PCT gene) and have done several experiments on it. | ||
| + | From our experiment, we use the optimized construct as one of the parts of our PLA production system. | ||
| + | <h2>Validation of Bioplastic Production</h2> | ||
| + | <p>In order to observe the conversion performance, infrared spectroscopic analysis was applied since PHA and PLA have their particular functional group in chemical structure, and are performed in particular wavelengths. The absorption peak of PLA is at 1081, 1188, 1364, 1452 , 1751 cm<sup>-1[1]</sup>, whereas the absorption peak 979, 1057, 1100, 1282, 1723, 2934, 2977cm<sup>-1[2]</sup>. We have compared and analyse the peak of the wavelength to confirm the bioplastic product we have produced. | ||
| + | In Figure 2, no absorption peak of PHA and PLA was identified, which indicated that the vector (pETDuet Vector) cannot give the desired product. On the other hand, the absorption peaks of PHA and PLA were identified after incubation, which suggested that the BBa_K3863004 and BBa_K3863008, the bioplastic performs significantly in the transformation process. Furthermore, the absorption peaks of PHA were found, which showed that BBa_K3863007 performs well and was the product that we expected to have. These results have proven that the bioplastic that we have produced can be formed significantly with BBa_K3863004, BBa_K3863008 and BBa_K3863007. | ||
| + | </p> | ||
| + | <img style="width:100%; height:auto" src="https://2021.igem.org/wiki/images/7/75/T--PuiChing_Macau--result13.jpg"> | ||
| + | <center><p>Figure 1a. IR spectrum of pETDuet vector</p></center> | ||
| + | <img style="width:100%; height:auto" src="https://2021.igem.org/wiki/images/e/ed/T--PuiChing_Macau--result14.jpg"> | ||
| + | <center><p>Figure 1b. IR spectrum of BBa_K3863004 </p></center> | ||
| − | + | <h2>Quantitative Measure of Bioplastic Production</h2> | |
| − | + | <p>In order to measure the amount of bioplastic produced, we used 20 microgram Nile Red (sigma aldrich N3013) per milliliter of agar to make a Nile agar plate and streak the cell culture (BBa_K3863007(PHA), BBa_K3863004(PCT) , BBa_K3863008(Phasin)) on the plate. After incubating for 2 days at 37 celsius in darkness. Stereomicroscope (Nikon SMZ18) is used to measure the intensity of Nile red. </p> | |
| + | <img style="width:100%; height:auto" src="https://2021.igem.org/wiki/images/c/c5/T--PuiChing_Macau--result0000.jpg"> | ||
| + | <center><p>Fig 3.Mean intensity of Nile red for quantitative measure of bioplastic production of (BBa_K3863007(PHA), BBa_K3863004(PhaC), BBa_K3863008(Phasin))</p></center> | ||
| + | <h2>Reference</h2> | ||
| + | <p>[1]https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3450024/pdf/12088_2009_Article_31.pdf<br> | ||
| + | [2]https://www.researchgate.net/figure/Fourier-transform-infrared-FTIR-spectra-of-PLA-PEG-PLA-PEG-blend-and-PLA-PEG-xGnP_fig1_277675367</p> | ||
| + | </html> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| − | + | <h2>Usage and Biology</h2> | |
<!-- --> | <!-- --> | ||
| Line 63: | Line 86: | ||
<partinfo>BBa_K1211001 parameters</partinfo> | <partinfo>BBa_K1211001 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | == Contribution == | ||
| + | *'''Group:''' iGEM Evry 2016 | ||
| + | *'''Author:''' Toky RATOVOMANANA | ||
| + | *'''Summary:''' We codon optimize the part for Pseudomonas putida, which is a safe organism reported to be efficient for polymerization. The new biobrick <html><a href="https://parts.igem.org/Part:BBa_K2042000">BBa_K2042000</a></html>, contain also a variant amino acid substitution (A243T) in order to produce (D)-Lactyl-CoA. | ||
Latest revision as of 15:52, 21 October 2021
Clostridum propionicum propionate CoA transferase
This is the DNA coding for the enzyme propionate CoA transferase from Clostridum propionicum. The enzyme normally catalyzes the reaction: acetyl-CoA + propanoate <--> acetate + propanoyl-CoA. This mutated sequence has four point mutations and one amino acid substitution (compared to the wild type enzyme) in order to produce (D)-Lactyl-CoA.
We used this enzyme as a the first enzyme in the pathway to create PLA or Polylactic acid. However, this biobrick produces (D)-Lactyl-CoA, which is the precursor to all PHAs, and thus this biobrick can be used in a wide range of projects in the future.
Data
Here is a gel showing our assembly of this gene. In order project we attached a promoter and terminator to the gene as well.


- Our project used this gene along with a Pseudomonas resinovorans PHA synthase gene to produce PLA. However, other teams can add other heterologous enzymes and produce different PHA. We tested the ability of our E. coli to produce PLA by using Nile red
- Nile red is an intercellular lipid strain
- Nile red does not affect the growth of bacteria, and its fluorescence is quenched in water
- We then proceeded to test our strain with out plasmid in the plate reader.
- Cells were grown for 24 hours with both enzymes induced and in the presence of Nile red. The cells were washed and re-suspended in PBS. The readings were normalized for optical density. Clearly, the one with our plasmid is more fluorescent indicating PLA production.

- After creating diversity using MAGE or Multiplex automated genome engineering, we needed a way to sort out those with the highest levels of fluorescence indicating greater PLA production. We decided to use FACS or FLuorescence activated cell sorting, to select those cells with the highest levels of fluorescence.
To read more about our project [http://2013.igem.org/Team:Yale/Project_Overview click here]

Team PuiChing Macau 2021: Optimization and Plastic Production
Based on the previous construct of Yale 2013 (BBa_K1211001), we have optimized the sequence(the PCT gene) and have done several experiments on it. From our experiment, we use the optimized construct as one of the parts of our PLA production system.Validation of Bioplastic Production
In order to observe the conversion performance, infrared spectroscopic analysis was applied since PHA and PLA have their particular functional group in chemical structure, and are performed in particular wavelengths. The absorption peak of PLA is at 1081, 1188, 1364, 1452 , 1751 cm-1[1], whereas the absorption peak 979, 1057, 1100, 1282, 1723, 2934, 2977cm-1[2]. We have compared and analyse the peak of the wavelength to confirm the bioplastic product we have produced. In Figure 2, no absorption peak of PHA and PLA was identified, which indicated that the vector (pETDuet Vector) cannot give the desired product. On the other hand, the absorption peaks of PHA and PLA were identified after incubation, which suggested that the BBa_K3863004 and BBa_K3863008, the bioplastic performs significantly in the transformation process. Furthermore, the absorption peaks of PHA were found, which showed that BBa_K3863007 performs well and was the product that we expected to have. These results have proven that the bioplastic that we have produced can be formed significantly with BBa_K3863004, BBa_K3863008 and BBa_K3863007.

Figure 1a. IR spectrum of pETDuet vector

Figure 1b. IR spectrum of BBa_K3863004
Quantitative Measure of Bioplastic Production
In order to measure the amount of bioplastic produced, we used 20 microgram Nile Red (sigma aldrich N3013) per milliliter of agar to make a Nile agar plate and streak the cell culture (BBa_K3863007(PHA), BBa_K3863004(PCT) , BBa_K3863008(Phasin)) on the plate. After incubating for 2 days at 37 celsius in darkness. Stereomicroscope (Nikon SMZ18) is used to measure the intensity of Nile red.
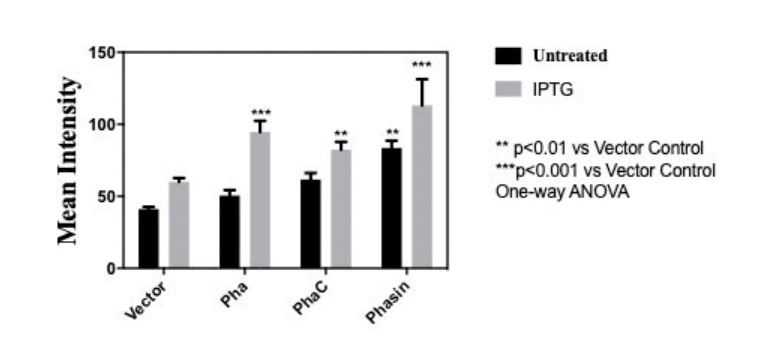
Fig 3.Mean intensity of Nile red for quantitative measure of bioplastic production of (BBa_K3863007(PHA), BBa_K3863004(PhaC), BBa_K3863008(Phasin))
Reference
[1]https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3450024/pdf/12088_2009_Article_31.pdf
[2]https://www.researchgate.net/figure/Fourier-transform-infrared-FTIR-spectra-of-PLA-PEG-PLA-PEG-blend-and-PLA-PEG-xGnP_fig1_277675367
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Contribution
- Group: iGEM Evry 2016
- Author: Toky RATOVOMANANA
- Summary: We codon optimize the part for Pseudomonas putida, which is a safe organism reported to be efficient for polymerization. The new biobrick BBa_K2042000, contain also a variant amino acid substitution (A243T) in order to produce (D)-Lactyl-CoA.



