Difference between revisions of "Part:BBa K1189000"
| (3 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1189000 short</partinfo> | <partinfo>BBa_K1189000 short</partinfo> | ||
| + | <html> | ||
| + | <p> | ||
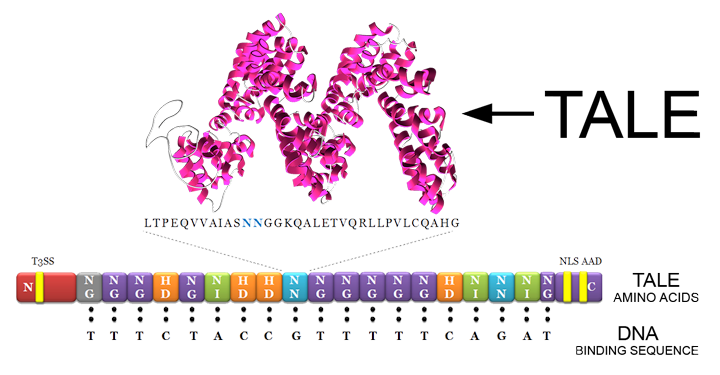
| + | Transcriptor Activator-Like Effectors (TALEs) are proteins produced by bacteria of the genus Xanthomonas and secreted into plant cells. These naturally occurring TALEs play a key role in bacterial infection, as they are responsible for up regulation of the host genes required for pathogenic growth and expansion (Mussolino and Cathomen, 2012). These special bacterial plant pathogen proteins bind to DNA by specifically recognizing one base pair with a single tandem repeat in their DNA-binding domain. TALEs are an advantageous tool in synthetic biology because they can be modified to bind to a chosen DNA sequence. | ||
| + | The central region of the protein, also termed repeat region, mediates DNA recognition through tandem repeats of 33 to 35 amino acids residues each (Bogdanove <i>et al.</i>, 2010). The binding domain usually comprises 15.5 to 19.5 single repeats. The last repeat, close to the C-terminus, is called “half-repeat” because it is only approximately 20 amino acids in length. Although the modules have conserved sequences, polymorphisms are found in residues 12 and 13, the “repeat-variable di-residue” (RVD). RVDs are specific for a single nucleotide; therefore, 19.5 repeat units target a specific 20-nucleotide sequence in the DNA (Mussolino and Cathomen, 2012). | ||
| + | </p> | ||
| + | <p> | ||
| + | TALE A was originally designed by Slovenia's 2012 iGEM team (<a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K782004"><b>BBa_K782004</b></a>). However, Slovenia's team made the part for use in eukaryotic cells. We modified the part so that it can be used in <i>E. coli</i> by removing the Kozak sequence and nuclear localization sequence, and codon optimizing it for <i>E. coli</i>. </p> | ||
| + | |||
| + | </html> | ||
| − | |||
<html> | <html> | ||
<figure> | <figure> | ||
| Line 11: | Line 19: | ||
[[File:Calgary2013-TALE-RVD.png]] | [[File:Calgary2013-TALE-RVD.png]] | ||
| − | <html><figcaption> | + | <html><figcaption>Figure 1: Shows the repeat variable domain(RVD) of TALEs. These RVDs are very specific and the code of 2 amino acids that bind to each nucleotide has been solved making TALEs very easy to engineer and very specific to their targets</figcaption> |
Latest revision as of 01:34, 26 October 2013
TALE-A with a his tag under a lacI promoter
Transcriptor Activator-Like Effectors (TALEs) are proteins produced by bacteria of the genus Xanthomonas and secreted into plant cells. These naturally occurring TALEs play a key role in bacterial infection, as they are responsible for up regulation of the host genes required for pathogenic growth and expansion (Mussolino and Cathomen, 2012). These special bacterial plant pathogen proteins bind to DNA by specifically recognizing one base pair with a single tandem repeat in their DNA-binding domain. TALEs are an advantageous tool in synthetic biology because they can be modified to bind to a chosen DNA sequence. The central region of the protein, also termed repeat region, mediates DNA recognition through tandem repeats of 33 to 35 amino acids residues each (Bogdanove et al., 2010). The binding domain usually comprises 15.5 to 19.5 single repeats. The last repeat, close to the C-terminus, is called “half-repeat” because it is only approximately 20 amino acids in length. Although the modules have conserved sequences, polymorphisms are found in residues 12 and 13, the “repeat-variable di-residue” (RVD). RVDs are specific for a single nucleotide; therefore, 19.5 repeat units target a specific 20-nucleotide sequence in the DNA (Mussolino and Cathomen, 2012).
TALE A was originally designed by Slovenia's 2012 iGEM team (BBa_K782004). However, Slovenia's team made the part for use in eukaryotic cells. We modified the part so that it can be used in E. coli by removing the Kozak sequence and nuclear localization sequence, and codon optimizing it for E. coli.

Figure 6: TALE capture assay was done with TALE B (
BBa_K1189001
)and TALE A B-lac fusion (
BBa_K1189031
) and DNA (
BBa_K1189006
)
with both target sequences. If capture is successful, the B-lac is present in the well giving a colour change from pink to yellow when subjected to benzylpenicillin substrate solution within 20 minutes. The only wells that change colour are the first four wells which contain TALE B, specific DNA, and TALE A B-lac fusion and our positive control wells which are the controls for our fusion TALE A B-lac protein and our positive recombinant b-lac. All our other controls including our test using a non-specific sequence of DNA remained pink . This preliminary characterization data demonstrates that the TALEs are able to bind to DNA with specificity. Additionally it also shows that our system of capturing DNA with two detector TALEs and then subsequent reporting of the DNA’s presence works.

This assay shows that we can capture our target DNA with two detector TALEs with specificity . Additionally, we can report whether that DNA has been captured and is present in the sample, which is a very important concept for our sensor system.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 2654
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]