Difference between revisions of "Part:BBa K1152013"
m (→Improved Production of Indigoidine by Co-transformation of Host Strain with Supplementary PPTases) |
|||
| (28 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
| − | + | [[file:iGEM_HD13_kuvette.png|300px|thumb|right|'''Figure 1''': Indigoidine Production in ''E. coli'' liquid cultures]] | |
| + | The IndC Indigoidine Synthetase from Photorhabdus luminescens laumondii TT01 is a nonribosomal peptide synthetase (NRPS) which produces the blue pigment indigoidine. Indigoidine consists of two cyclic glutamines. IndC is a single module NRPS and consists of an Adenylation-, Thiolation- and a Thioesterase-domain. The A-domain thus contains an internal Oxidation domain. | ||
| + | |||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | ||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K1152013 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1152013 SequenceAndFeatures</partinfo> | ||
| − | + | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
| Line 17: | Line 20: | ||
<partinfo>BBa_K1152013 parameters</partinfo> | <partinfo>BBa_K1152013 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | ===Expression of functional indigoidine synthetase indC derived from ''P. luminescens'' in five substrains of ''E. coli''=== | ||
| + | <br> | ||
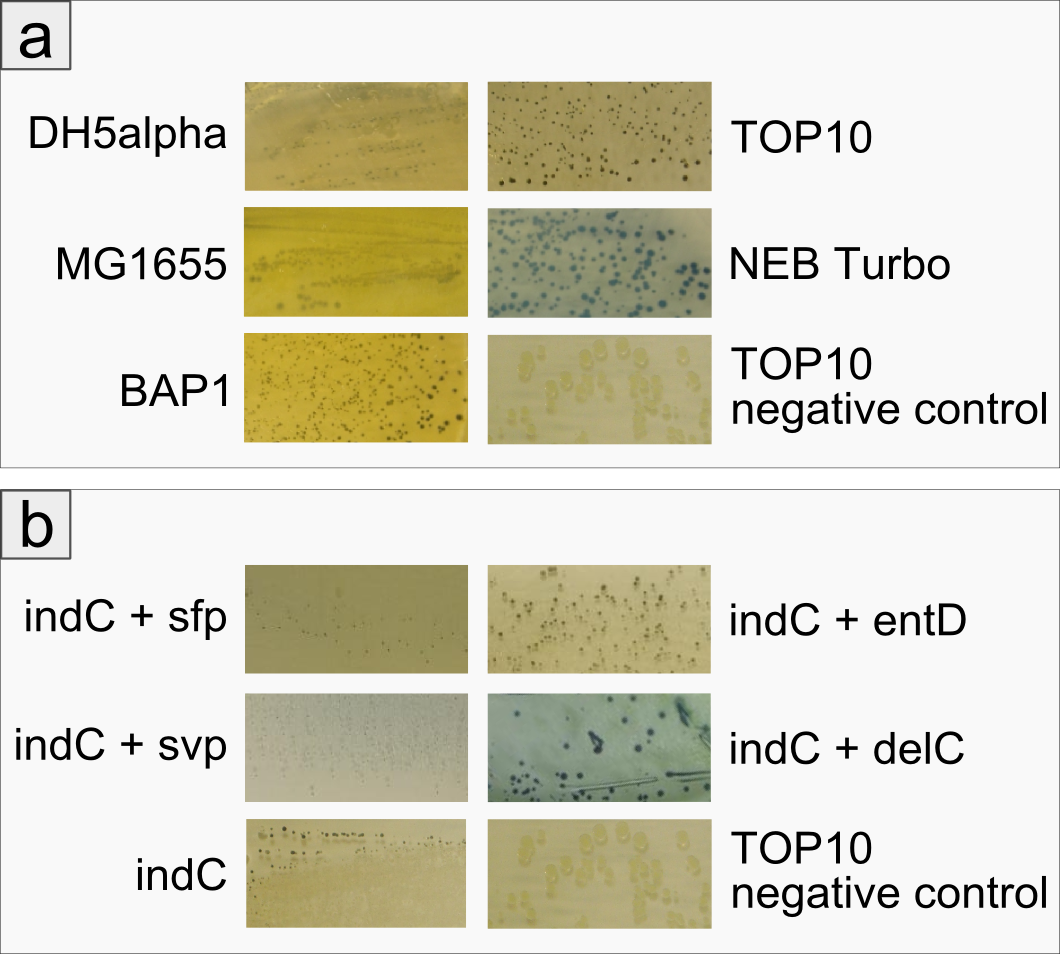
| + | The open reading frame of the native indigoidine synthetase indC was amplified from genomic DNA of ''P. lumninescens laumondii'' TT01 and cloned into a plasmid under the control of an lac-inducible promoter. This indC expression cassette was transformed into different substrains of ''E. coli'', namely DH5alpha, MG1655, BAP1, TOP10 and NEB Turbo. All of these host strains express the endogenous PPTase entD which is responsible for the transfer of the 4'-phosphopantetheine residue from coenzyme A to the T-domain of EntF, a NRPS module involved in the enterobactin pathway (Lambalot 1996). As depicted in Figure 2a, entD does not exhibit strict substrate specificity being restricted to activating domains of the enterobactin pathway, but is able to activate the T-domain of indC as determined by the blue phenotype of the transformed cells. Except for NEB Turbo cells, all transformed host strains displayed a decellerated growth and significantly smaller colonies on plate when compared to the negative control. NEB Turbo showed regular colony growth and developed a strong blue phenotype upon induction with IPTG. All host strains were able to express the functional indigoidine sythetase. | ||
| + | |||
| + | ===Improved Production of Indigoidine by Co-transformation of Host Strain with Supplementary PPTases=== | ||
| + | The expression of indC under activation by the endogenous PPTase entD was sufficient for easy detection of indigoidine production on plates harboring indC-carrying ''E. coli'' cells. In order to determine, whether the amount of indigoidine production in the ''E. coli''TOP10 cells is dependent on the quality of the interaction of indC with the PPTase, four PPTases derived from varying origins were selected and amplified from the genome of the hosts of origin. ''E. coli'' TOP10 cells were co-transformed with plasmids coding for the different PPTases and the plasmid containing the expression cassette for indC. As reference for the endogenous PPTase activity served cells only transformed with the indC plasmid. Irrespective of the PPTase, growth of colonies was retarded. Remarkably however, colonies co-transformed with the PPTase plasmid remained of smaller size than the ones only carrying the indC construct. On the other side, indigoidine production was more diffuse in the latter cells with secretion of the blue pigment into the agar (Figure 2b, indC) and only slight blue-greenish coloring of the colonies. The four PPTases additionally introduced into the TOP10 cells were all shown to be functional (blue phenotype of the transformants, Figure 6b), but lead to the retention of most of the indigoidine within the cells. Colonies of cells transformed with thess constructs, were of convex shape and of distinct, dark blue color. Overall, cells carrying an additional PPTase showed increased indigoidine production compared to the cells relying on the endogenous entD. | ||
| + | |||
| + | |||
| + | [[image:Heidelberg_IndPD_Fig6.png|400px]] | ||
| + | |||
| + | '''Figure 2''': Comparison between different E. coli strains and PPTases: a) Comparison of different E. coli strains examining growth and indigoidine production The figure shows five different strains of E. coli that have been co-transformed with an indC expression plasmid and a sfp expression plasmid. The negative control is E. coli TOP10 without a plasmid. All transformants have been grown on LB agar for 48 hours at room temperature, cells were not induced. One can see that even without induction all strains express the indigoidine synthetase and produce the blue pigment indigoidine. However, the strains BAP1 and NEB Turbo grow faster in the first day, exhibiting a white phenotype (data not shown). Colonies on the plate of E. coli TOP10 are very small and dark blue/ black. Assuming that indigoidine production inhibits cell growth due to its toxicity, we concluded that TOP10 produced the most indigoidine among the strains we tested. We used E. coli TOP10 for the following experiments. b) Comparison between different PPTases concerning overall indigoidine production The Figure shows ''E. coli'' TOP10 cells co-transformed with indC and four different PPTases (sfp, svp, entD and delC), respectively. The image bottom left shows ''E. coli'' TOP10 cells without additional PPTase and the negative control is TOP10 without a plasmid. | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | Figure 3 shows the analytical restiction digest we performed for part validation: | ||
| + | [[File:Heidelberg2013_BBa_testdigest_indigoidine.png|280px|thumb|center|'''Figure 3:''' Restriction digest with EcoRI, XbaI, SpeI, PstI.]] | ||
| − | + | ===Source=== | |
| + | IndC was amplified from ''P. luminescens laumondii'' TT01 (DSM15139) genomic DNA. | ||
| + | <br> | ||
| + | <br> | ||
| + | ===References=== | ||
| + | #Duchaud, E. et. al., Nat. Biotechnol. 21 (11), 1307-1313 (2003): The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens | ||
| + | #Brachmann AO et. al. (2012) Triggering the production of the cryptic blue pigment indigoidine from Photorhabdus luminescens. J Biotechnol 157: 96-99. | ||
| + | #Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT (1996) A new enzyme superfamily - the phosphopantetheinyl transferases. Chem Biol 3: 923-936. | ||
Latest revision as of 18:47, 6 October 2013
IndC Indigoidine Synthetase device
The IndC Indigoidine Synthetase from Photorhabdus luminescens laumondii TT01 is a nonribosomal peptide synthetase (NRPS) which produces the blue pigment indigoidine. Indigoidine consists of two cyclic glutamines. IndC is a single module NRPS and consists of an Adenylation-, Thiolation- and a Thioesterase-domain. The A-domain thus contains an internal Oxidation domain.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 4087
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1467
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 2730
Expression of functional indigoidine synthetase indC derived from P. luminescens in five substrains of E. coli
The open reading frame of the native indigoidine synthetase indC was amplified from genomic DNA of P. lumninescens laumondii TT01 and cloned into a plasmid under the control of an lac-inducible promoter. This indC expression cassette was transformed into different substrains of E. coli, namely DH5alpha, MG1655, BAP1, TOP10 and NEB Turbo. All of these host strains express the endogenous PPTase entD which is responsible for the transfer of the 4'-phosphopantetheine residue from coenzyme A to the T-domain of EntF, a NRPS module involved in the enterobactin pathway (Lambalot 1996). As depicted in Figure 2a, entD does not exhibit strict substrate specificity being restricted to activating domains of the enterobactin pathway, but is able to activate the T-domain of indC as determined by the blue phenotype of the transformed cells. Except for NEB Turbo cells, all transformed host strains displayed a decellerated growth and significantly smaller colonies on plate when compared to the negative control. NEB Turbo showed regular colony growth and developed a strong blue phenotype upon induction with IPTG. All host strains were able to express the functional indigoidine sythetase.
Improved Production of Indigoidine by Co-transformation of Host Strain with Supplementary PPTases
The expression of indC under activation by the endogenous PPTase entD was sufficient for easy detection of indigoidine production on plates harboring indC-carrying E. coli cells. In order to determine, whether the amount of indigoidine production in the E. coliTOP10 cells is dependent on the quality of the interaction of indC with the PPTase, four PPTases derived from varying origins were selected and amplified from the genome of the hosts of origin. E. coli TOP10 cells were co-transformed with plasmids coding for the different PPTases and the plasmid containing the expression cassette for indC. As reference for the endogenous PPTase activity served cells only transformed with the indC plasmid. Irrespective of the PPTase, growth of colonies was retarded. Remarkably however, colonies co-transformed with the PPTase plasmid remained of smaller size than the ones only carrying the indC construct. On the other side, indigoidine production was more diffuse in the latter cells with secretion of the blue pigment into the agar (Figure 2b, indC) and only slight blue-greenish coloring of the colonies. The four PPTases additionally introduced into the TOP10 cells were all shown to be functional (blue phenotype of the transformants, Figure 6b), but lead to the retention of most of the indigoidine within the cells. Colonies of cells transformed with thess constructs, were of convex shape and of distinct, dark blue color. Overall, cells carrying an additional PPTase showed increased indigoidine production compared to the cells relying on the endogenous entD.
Figure 2: Comparison between different E. coli strains and PPTases: a) Comparison of different E. coli strains examining growth and indigoidine production The figure shows five different strains of E. coli that have been co-transformed with an indC expression plasmid and a sfp expression plasmid. The negative control is E. coli TOP10 without a plasmid. All transformants have been grown on LB agar for 48 hours at room temperature, cells were not induced. One can see that even without induction all strains express the indigoidine synthetase and produce the blue pigment indigoidine. However, the strains BAP1 and NEB Turbo grow faster in the first day, exhibiting a white phenotype (data not shown). Colonies on the plate of E. coli TOP10 are very small and dark blue/ black. Assuming that indigoidine production inhibits cell growth due to its toxicity, we concluded that TOP10 produced the most indigoidine among the strains we tested. We used E. coli TOP10 for the following experiments. b) Comparison between different PPTases concerning overall indigoidine production The Figure shows E. coli TOP10 cells co-transformed with indC and four different PPTases (sfp, svp, entD and delC), respectively. The image bottom left shows E. coli TOP10 cells without additional PPTase and the negative control is TOP10 without a plasmid.
Figure 3 shows the analytical restiction digest we performed for part validation:
Source
IndC was amplified from P. luminescens laumondii TT01 (DSM15139) genomic DNA.
References
- Duchaud, E. et. al., Nat. Biotechnol. 21 (11), 1307-1313 (2003): The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens
- Brachmann AO et. al. (2012) Triggering the production of the cryptic blue pigment indigoidine from Photorhabdus luminescens. J Biotechnol 157: 96-99.
- Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT (1996) A new enzyme superfamily - the phosphopantetheinyl transferases. Chem Biol 3: 923-936.