Difference between revisions of "Part:BBa K1022100"
(→Improvement by Team IISER_Kolkata 2021) |
|||
| (14 intermediate revisions by 3 users not shown) | |||
| Line 53: | Line 53: | ||
5. Cyflogic 1.2.1., available on:http://www.cyflogic.com/. | 5. Cyflogic 1.2.1., available on:http://www.cyflogic.com/. | ||
| + | <h2> <b> University of Nebraska-Lincoln 2019 </b> </h2> | ||
| + | To characterize BBa_K3191101, cell cultures were grown alongside parts BBa_K1022100, BBa_K3191102, and BBa_K206000 cells. Cells were grown overnight and diluted to an OD of 0.05 in the morning with LB. The cultures were then allowed to grow to an OD 0D of 0.5. At this point, cells were washed with PBS and resuspended with 120 mM, pH 8.0 Tris-HCl. 30 mM EDTA and 10 mM of arabinose was then added to the cultures and the cells were incubated for 2 minutes. The cultures were then diluted with 2 mL of LB and antibiotics, and the cells were placed in a 37C shaker. Fluorescence was recorded every hour for three hours. | ||
| + | |||
| + | <br> https://2019.igem.org/wiki/images/0/07/T--UNebraska-Lincoln--007.jpg <br> | ||
| + | |||
| + | <p style="text-align:center">Figure 1: Total fluorescence of pre-permeabilized cells with and without AIP induction.</p> | ||
| + | |||
| + | <br> https://2019.igem.org/wiki/images/9/98/T--UNebraska-Lincoln--006.jpg <br> | ||
| + | |||
| + | <p style="text-align:center">Figure 2: Total fluorescence of permeabilized cells at 0 and 3 hours post-induction with AIP. </p> | ||
| + | |||
| + | <br> https://2019.igem.org/wiki/images/6/6e/T--UNebraska-Lincoln--009.jpg <br> | ||
| + | |||
| + | <p style="text-align:center">Figure 3: Total change in fluorescence of permeabilized cells with and without AIP induction. </p> | ||
| + | |||
| + | <h4> Results </h4> | ||
| + | <p> | ||
| + | Our characterization data displays the difference in fluorescence values between BBa_K206000, BBa_K1022100, BBa_K3191101, and BBa_K3191102. This data was analyzed with a two-tail t-test of unequal variances with a significance level of 0.05 to draw conclusions of significance. Based on these values, fluorescence for BBa_K1022100 and BBa_K3191101 tend to exceed the control. This demonstrates that BBa_K1022100 and BBa_K3191101 both function properly. | ||
| + | </p> | ||
| + | <p> | ||
| + | Figure 1 demonstrates fluorescence values under the pre-permeabilization protocol. These data indicate that AIP induction caused minimal difference in fluorescence intensity. Figures 2 and 3 show fluorescence intensity and change in fluorescence, respectively, of cells after the permeabilization protocol. When comparing total fluorescence, BBa_K1022100 tends to yield significantly greater fluorescence values than BBa_K3191101 at hour 8, as supported by a p-values of 0.002 for the AIP trial. This suggests the modified sequences, while more complete, do not successfully increase GFP production under the original characterization procedures. The addition of SarA also does not appear to provide a significantly greater fluorescence under the influence of AIP. This is seen in a comparison of BBa_K3191101 and BBa_K3191102 at hour 3 in figure 4 (p-value = 0.902). | ||
| + | </p> | ||
| + | <p> | ||
| + | Trends in the data suggest that the introduction of AIP appears to have a greater effect on BBa_K3191101 and BBa_K3191102 than on BBa_K1022100 under the permeabilization characterization conditions. The fluorescence values for these conditions are seen in figures 2 and 3. In these conditions, a Tris-EDTA solution is used to attempt to increase AIP intake by the cells. BBa_K1022100 does not experience significantly greater fluorescence than the control at hour 3 with AIP induction (p-value = 0.111). In comparison, BBa_K3191101 and BBa_K3191102 have significantly greater fluorescence at hour 3 with AIP induction. This is demonstrated with p-values of 0.010 for BBa_K3191101 and 0.002 for BBa_K3191102. This suggests that AIP induction does not significantly increase fluorescence in BBa_K1022100, while it does increase in BBa_K3191101 and BBa_K3191102. | ||
| + | </p> | ||
| + | <p> | ||
| + | The increase in fluorescence may be explained by the modified experimental procedure. If the presence of Tris-EDTA successfully increases AIP uptake, then an increase in AIP uptake only improves fluorescence in BBa_K3191101 and BBa_K3191102. This suggests that our parts are truly induced by AIP, while BBa_K1022100 is not. | ||
| + | |||
| + | =='''Improvement by Team IISER_Kolkata 2021'''== | ||
| + | '''Group:''' iGEM21_IISER_Kolkata <br> | ||
| + | '''Author:''' Debdeep Chatterjee, Shubhamay Das <br> | ||
| + | '''Summary:''' Our team made an improvement of this exciting part(BBa_K1022100) and created an improved part named Bidirectional AIP-1 sensor(BBa_K3799057). Below is the description of our improved part.<br> | ||
| + | |||
| + | '''Bidirectional AIP-1 sensor'''<br> | ||
| + | This is a high throughput sensing module that can sense the presence of AIP-I molecules. This part consists of a pBAD promoter, an AIP sensor infrastructure (BBa_I746101), an AIP inducible promoter P2 and a GFP reporter. | ||
| + | |||
| + | In the natural system, the signalling Auto-inducing peptide (termed AIP) is made from AgrD, while AgrB, a transmembrane protein that is responsible for the cyclization. AgrC and AgrA, form the two-component system responsible for AIP detection and response relay, respectively. Upon binding to AIP, AgrC, a transmembrane histidine kinase receptor, facilitates the phosphorylation of the transcriptional activator AgrA. The phosphorylated AgrA, in turn, activates gene expression regulated by P2 and P3 promoters. There are four known variants of AIP (AIP I-IV) with different molecular structures and cross-inhibitory activity. | ||
| + | |||
| + | This part has been improved from [https://parts.igem.org/Part:BBa_K1022100 BBa_K1022100] by flipping the direction of p2 promoter to the opposite direction of pBAD promoter. | ||
| + | |||
| + | In the existing part '''(Unidirectional circuit)''', we observed a significant increase in GFP expression in samples with zero AIP concentration. Based on this observation, we inferred that this could be due to the leaky gene expression of GFP. We have improved the existing part in such a way that the leaky gene expression reduced significantly. | ||
| + | |||
| + | '''Design:'''<br> | ||
| + | To solve the leaky gene expression we designed a '''bidirectional circuit'''. We Used Hi-Fi DNA assembly kit to make this bidirectional circuit. First, we divided the existing part (BBa_K1022100) into three segments and ordered those from Twist Bioscience. These three fragments were assembled in two different ways to produce the existing part as well as our designed improved part. For these assemblies, we designed two different sets of primers (a total of eight primers) using the NEBilder tool. We extended our three gene fragments with appropriate primers to produce overlap sequences at both ends of each fragment. | ||
| + | |||
| + | To make the improved part, we designed the overlap sequences in such a way that the p2 promoter along with the GFP reporter is flipped. This will make the direction of pBAD promoter and P2 promoter opposite to each other, which will significantly decrease the leaky gene expression of the GFP reporter. | ||
| + | |||
| + | '''-----Charaterisation-----'''<br> | ||
| + | '''Introduction''' | ||
| + | |||
| + | We observed that there is leaky gene expression in the Unidirectional circuit. We modified the existing part to reduce the leaky gene expression. By flipping the gene we expect that the leaky gene expression will be reduced. To check our hypothesis, we have compared the expression of GFP in the unidirectional and the bidirectional genetic circuit. | ||
| + | |||
| + | '''Experiment''' | ||
| + | |||
| + | First, we PCR extended the three DNA segments (pBAD-agrC, P2-GFP,agrA,) of the unidirectional circuit with specific primers. To confirm the success of PCR extension an Agarose Gel Electrophoresis was performed. As evident from the picture below, correct bands for all three amplified DNA segments were obtained. (Extended pBAD-agrC ~ 1.4 Kb, Extended P2-GFP ~ 1Kb, Extended agrA ~ 800bp and extended backbone~2kb. | ||
| + | [[Image:T--IISER_Kolkata--uni_colony_pcr.jpg | thumb | center | 700 px |PCR extension of pBAD-agrC,P2-GFP,agrA for unidirectional assembly | ||
| + | ]] | ||
| + | [[Image:T--IISER_Kolkata--psb1c3.jpeg | thumb | center | 800 px |PCR extension of psb1C3 for unidirectional assembly]] | ||
| + | |||
| + | After that, the same four segments (pBAD-agrC, P2-GFP, agrA, and psb1C3 plasmid backbone) were PCR amplified with different sets of specific primers. This extended overlap helps flip the P2-GFP fragments. From the gel electrophoresis image below, we can observe similar band sizes as above because the difference in length of the extension is very low. | ||
| + | [[Image:T--IISER_Kolkata--bidi_extension.jpg | thumb | center | 700 px |PCR extension of pBAD-agrC,P2-GFP,agrA for bidirectional assembly | ||
| + | ]] | ||
| + | [[Image:T--IISER_Kolkata--AgrA_extension.jpg | thumb | center | 700 px | PCR extention of agrA segment | ||
| + | ]] | ||
| + | After the successful extension of all the segments, those extended fragments were assembled using Hi-Fi DNA assembly master mix. The reaction mixture was transformed in the E.Coil DH5a. After that, colony PCR of both the unidirectional and bidirectional circuit was performed using VF2 and VR primers to identify the bacterial colony with our desired gene circuits. | ||
| + | |||
| + | As evident from the colony PCR results, positive inserts for both unidirectional and bidirectional assemblies were obtained. | ||
| + | [[Image:T--IISER_Kolkata--uni_pcr_colony.jpg | thumb | center | 700 px | Colony PCR result of unidirectional assembly with VF2 and VR | ||
| + | ]] | ||
| + | [[Image:T--IISER_Kolkata--bidicolonypcr.jpg | thumb | center | 700 px | Colony PCR result of bidirectional assembly with VF2 and VR]] | ||
| + | |||
| + | |||
| + | After the successful assembling of both the fragments, we planned to see the expression level of GFP in both bidirectional and unidirectional circuits. For that, first the AIP-1 generator (BBa_K3799006) was expressed in 40ml of bacterial culture to obtain AIP-1 molecules. After expressing for 8-10 hours, the culture was centrifuged and the supernatant was filter-sterilized. The supernatant was lyophilized to concentrate the AIP-1 molecule. After full lyophilization, it was resuspended in 3 ml of NFW to make a stock solution of AIP1. | ||
| + | |||
| + | The stock solution of AIP-1 molecules was diluted to make solutions of different concentrations, from 0% to 100%, where 0% was the negative control and 100% was the one-third dilution of our stock solution. | ||
| + | |||
| + | The two types of cells were incubated (one with unidirectional plasmid and the other with bidirectional plasmid) at 30℃ for 8 hours without AIP-1. | ||
| + | |||
| + | The fluorescence intensity of those two types of cells in absence of AIP-1 molecules was measured at different time points and it was observed that there is a higher fluorescence intensity in cells with unidirectional circuits as compared to the cells with bidirectional circuits. This infers that the bidirectional circuit has less leaky gene expression than that of the unidirectional circuit. | ||
| + | |||
| + | [[Image:IISER_Kolkata--Bi-Intensity_vs_time.jpeg | thumb | center | 800 px | Fluorescence assay of bidirectional and unidirectional circuits without AIP-1 vs Time]] | ||
| + | |||
| + | We also wanted to check whether this difference in leaky gene expression is consistent with different concentrations of AIP-1. | ||
| + | |||
| + | To check this, we induced our two types of cells (one with unidirectional plasmid and the other with bidirectional plasmid) with different concentrations of the AIP-1 solution that we mentioned above. From our data, we infer that this leaky gene expression of the unidirectional circuit is consistent in different concentrations of AIP-1 molecules. Also, we can observe a slight decrease in the gap of expression of GFP as the concentration of AIP-1 increases. This may be because of the fact that the ratio of leaky gene expression and the AIP-1 induced expression decreases as the AIP-1 concentration increases. | ||
| + | |||
| + | |||
| + | [[Image:IISER Kolkata--Bi-Intensity vs conc.jpeg | thumb | center | 800 px |Fluorescence assay of bidirectional and unidirectional circuits at different concentrations of AIP-1 at 5hrs ]] | ||
| + | |||
| + | '''Final Comments:''' | ||
| + | |||
| + | Thus we can conclude that our new part [https://parts.igem.org/Part:BBa_K3799057 BBa_K3799057] is indeed an improvement of the existing part [https://parts.igem.org/Part:BBa_K1022100 BBa_K1022100] in significantly decreasing the leaky gene expression under the P2 promoter | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
<partinfo>BBa_K1022100 parameters</partinfo> | <partinfo>BBa_K1022100 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
Latest revision as of 02:40, 22 October 2021
pBAD:AIP receiver:GFP:TT
This plasmid contains code for pBAD promoter, AIP receiver Agr A and Agr C, GFP along with double terminator.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 125
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 521
Illegal BamHI site found at 65
Illegal BamHI site found at 1405 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 2970
Characterization
For more info, visit [http://2013.igem.org/Team:TU-Delft/Sensing Sensing AIP]on TU Delft iGEM13 Wiki
Sensor Experiment
Introduction:
In this sensor test-construct AgrC and AgrA are expressed, after which AgrC can act as a receptor for AIP. After binding AIP, AgrC will phosphorylate AgrA which then starts acting as a postitive transcription factor for pP2[1]. Therefore, upon induction by AIP, the construct will give a GFP signal. This is tested using the following experiment.
Experimental Set Up:
Cells are induced with 0.1 % Arabinose (to induce pBAD) and AIP’s, and with only AIP. Induction with AIP is done at 1µM and 10 µM. 3 hours after induction fluorescence-activated cell sorting (FACS) is used to check for GFP signals. The controls are BL21(DE3) cells, uninduced plasmid containing BL21(DE3) and a constitutively expressed GFP construct containing strain. The AIP used to induce the cells was synthesized by Eurogentec.
The cells induced with arabinose all died, which could be seen by the fact the signal given by the FACS, this could be due the fact AgrA and/or AgrC are toxic in high concentrations. The fact E. coli is still able to detect AIPs without induction of the Agr genes is explainable due the fact pBAD is a bit leaky, making low transcription possible, apparently only a small amount of activated AgrA is required to give a measurable level of GFP.
Earlier work on the detection of AIPs by Gram-negatives has been done, but with unclear or non-convincing results [2,3]. Often the outer-membrane of E. coli was seen as an obstacle for the implementation of auto-inducing peptide sensing systems in Gram negatives. Although, after intensive literature research we found a molecule very similar to the AIP molecule used in these experiments, that is known to pass the outer membrane. The molecule bacitracin has been shown to interfere with cell wall synthesis in Gram negative species, therefore being able to pass the outer membrane as cell wall synthesis occurs in the periplasm. Comparison of the molecular structures of bacitracin and AIP regarding size, molecular weight, polarity and charge gave us an indication AIP should be possible to pass through the outer membrane of E. coli.
Results:
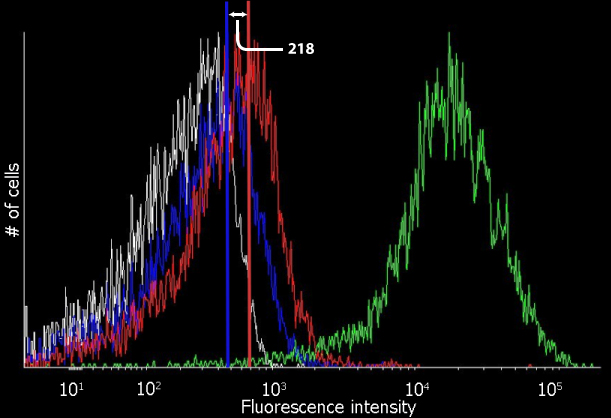
Using Cyflogic 1.2.1 software [5], FACS data was analysed, showing a clear induction of the AIP receiver GFP construct by AIP. The difference between the blue and the red line (218) indicates the difference in fluorescent signal of uninduced and induced cells. Untransformed Bl21(DE3) and constitutively expressed GFP were used as respectively negative and positive control. The significant difference between the uninduced and the induced samples is determined with a P-value >10^-4.
Fluorescent signal measured with the FACS. The white line refers to the control BL21(DE3) cells. In blue the uninduced pBAD AIP receiver GFP is shown. The red line represents the AIP induced receiver cells. The fluorescence intensity scale is logarithmic and the total number of cells counted per sample is 10,000.
Discussion:
Taking the results described above into account, we are able to sense the presence of a Gram positive species with the use of E. coli transformed with our AIP receiver GFP construct. To our knowledge, we are the first to show the interspecies communication between Gram negative and Gram positive bacterial species. In this case the sensing of the AIP of S. aureus by E. coli. For the final application described in the [http://2013.igem.org/Team:TU-Delft/ProjectOverview TU Delft iGEM13 Wiki], the level of activation of the pP2 promoter by AgrA might be too low. In order to make the sensing system able to activate a peptide production system capable of producing a concentration high enough to kill or inhibit the growth of S. aureus, an amplification circuit could be used.
References
1. Philip Mdowell, Zina Affas, et al.,Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus, Molecular Microbiology, Volume 41, Issue 2, pages 503–512, Jul 2001.
2. http://2007.igem.org/wiki/index.php/Cambridge
3. http://2012.igem.org/Team:HIT-Harbin
4. T.J. Pollock, L. Thorne, Mechanism of bacitracin resistance in gram-negative bacteria that synthesize exopolysaccharides, J Bacteriol ; 176(20): 6229–6237, Oct 1994
5. Cyflogic 1.2.1., available on:http://www.cyflogic.com/.
University of Nebraska-Lincoln 2019
To characterize BBa_K3191101, cell cultures were grown alongside parts BBa_K1022100, BBa_K3191102, and BBa_K206000 cells. Cells were grown overnight and diluted to an OD of 0.05 in the morning with LB. The cultures were then allowed to grow to an OD 0D of 0.5. At this point, cells were washed with PBS and resuspended with 120 mM, pH 8.0 Tris-HCl. 30 mM EDTA and 10 mM of arabinose was then added to the cultures and the cells were incubated for 2 minutes. The cultures were then diluted with 2 mL of LB and antibiotics, and the cells were placed in a 37C shaker. Fluorescence was recorded every hour for three hours.
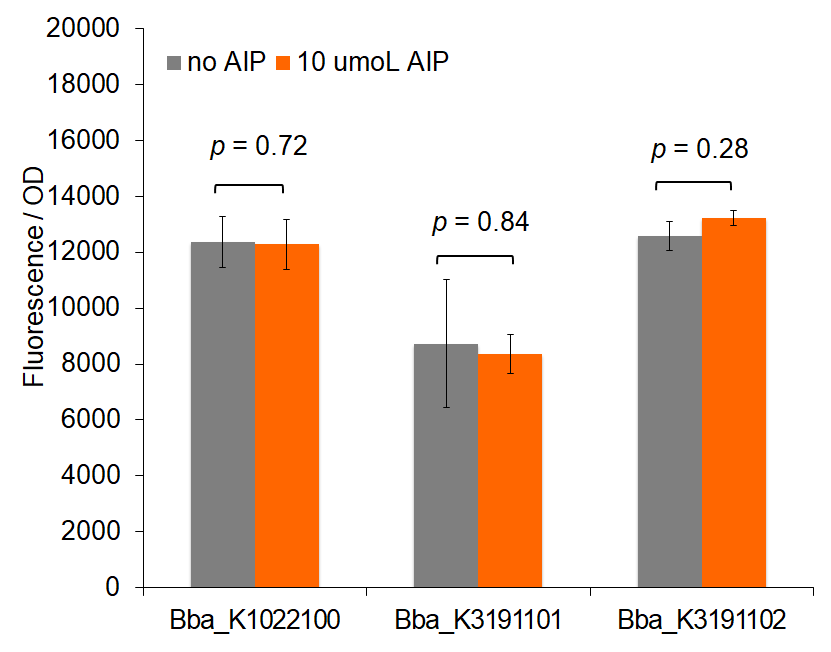
Figure 1: Total fluorescence of pre-permeabilized cells with and without AIP induction.
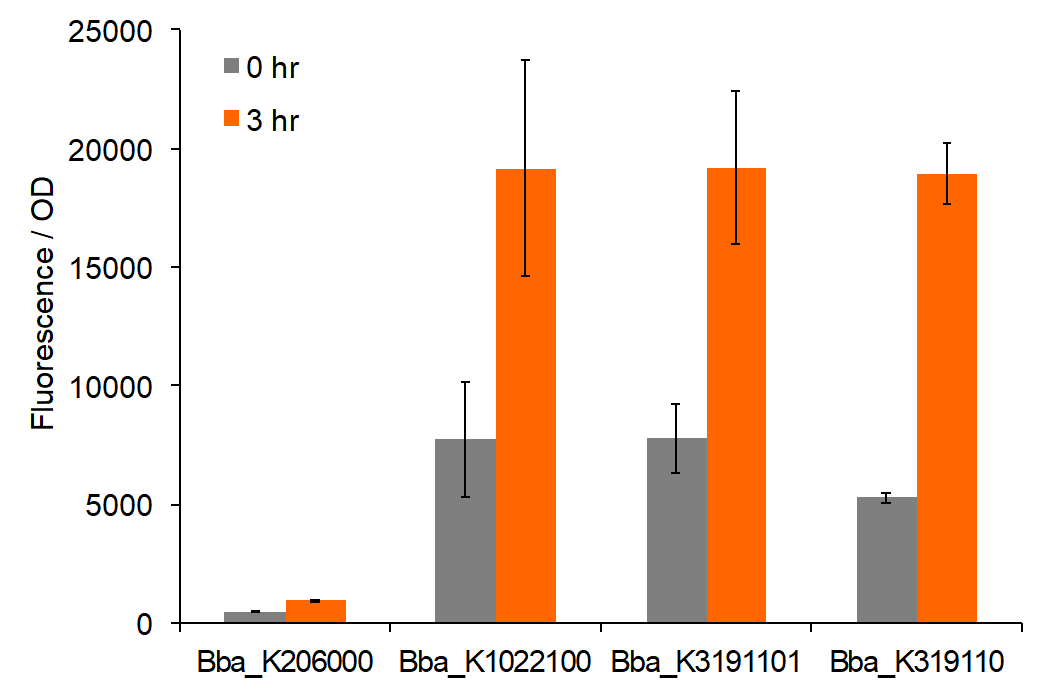
Figure 2: Total fluorescence of permeabilized cells at 0 and 3 hours post-induction with AIP.
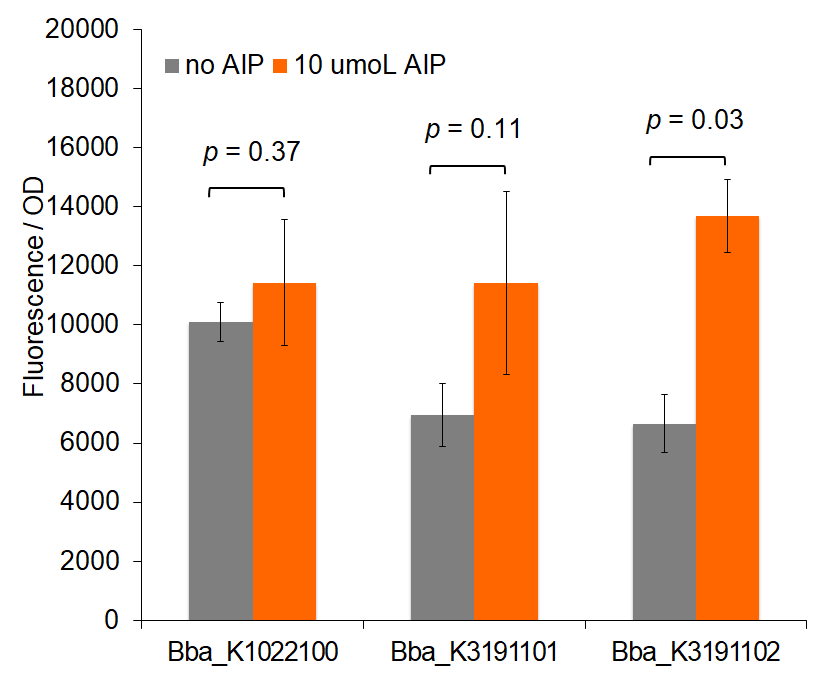
Figure 3: Total change in fluorescence of permeabilized cells with and without AIP induction.
Results
Our characterization data displays the difference in fluorescence values between BBa_K206000, BBa_K1022100, BBa_K3191101, and BBa_K3191102. This data was analyzed with a two-tail t-test of unequal variances with a significance level of 0.05 to draw conclusions of significance. Based on these values, fluorescence for BBa_K1022100 and BBa_K3191101 tend to exceed the control. This demonstrates that BBa_K1022100 and BBa_K3191101 both function properly.
Figure 1 demonstrates fluorescence values under the pre-permeabilization protocol. These data indicate that AIP induction caused minimal difference in fluorescence intensity. Figures 2 and 3 show fluorescence intensity and change in fluorescence, respectively, of cells after the permeabilization protocol. When comparing total fluorescence, BBa_K1022100 tends to yield significantly greater fluorescence values than BBa_K3191101 at hour 8, as supported by a p-values of 0.002 for the AIP trial. This suggests the modified sequences, while more complete, do not successfully increase GFP production under the original characterization procedures. The addition of SarA also does not appear to provide a significantly greater fluorescence under the influence of AIP. This is seen in a comparison of BBa_K3191101 and BBa_K3191102 at hour 3 in figure 4 (p-value = 0.902).
Trends in the data suggest that the introduction of AIP appears to have a greater effect on BBa_K3191101 and BBa_K3191102 than on BBa_K1022100 under the permeabilization characterization conditions. The fluorescence values for these conditions are seen in figures 2 and 3. In these conditions, a Tris-EDTA solution is used to attempt to increase AIP intake by the cells. BBa_K1022100 does not experience significantly greater fluorescence than the control at hour 3 with AIP induction (p-value = 0.111). In comparison, BBa_K3191101 and BBa_K3191102 have significantly greater fluorescence at hour 3 with AIP induction. This is demonstrated with p-values of 0.010 for BBa_K3191101 and 0.002 for BBa_K3191102. This suggests that AIP induction does not significantly increase fluorescence in BBa_K1022100, while it does increase in BBa_K3191101 and BBa_K3191102.
The increase in fluorescence may be explained by the modified experimental procedure. If the presence of Tris-EDTA successfully increases AIP uptake, then an increase in AIP uptake only improves fluorescence in BBa_K3191101 and BBa_K3191102. This suggests that our parts are truly induced by AIP, while BBa_K1022100 is not.
Improvement by Team IISER_Kolkata 2021
Group: iGEM21_IISER_Kolkata
Author: Debdeep Chatterjee, Shubhamay Das
Summary: Our team made an improvement of this exciting part(BBa_K1022100) and created an improved part named Bidirectional AIP-1 sensor(BBa_K3799057). Below is the description of our improved part.
Bidirectional AIP-1 sensor
This is a high throughput sensing module that can sense the presence of AIP-I molecules. This part consists of a pBAD promoter, an AIP sensor infrastructure (BBa_I746101), an AIP inducible promoter P2 and a GFP reporter.
In the natural system, the signalling Auto-inducing peptide (termed AIP) is made from AgrD, while AgrB, a transmembrane protein that is responsible for the cyclization. AgrC and AgrA, form the two-component system responsible for AIP detection and response relay, respectively. Upon binding to AIP, AgrC, a transmembrane histidine kinase receptor, facilitates the phosphorylation of the transcriptional activator AgrA. The phosphorylated AgrA, in turn, activates gene expression regulated by P2 and P3 promoters. There are four known variants of AIP (AIP I-IV) with different molecular structures and cross-inhibitory activity.
This part has been improved from BBa_K1022100 by flipping the direction of p2 promoter to the opposite direction of pBAD promoter.
In the existing part (Unidirectional circuit), we observed a significant increase in GFP expression in samples with zero AIP concentration. Based on this observation, we inferred that this could be due to the leaky gene expression of GFP. We have improved the existing part in such a way that the leaky gene expression reduced significantly.
Design:
To solve the leaky gene expression we designed a bidirectional circuit. We Used Hi-Fi DNA assembly kit to make this bidirectional circuit. First, we divided the existing part (BBa_K1022100) into three segments and ordered those from Twist Bioscience. These three fragments were assembled in two different ways to produce the existing part as well as our designed improved part. For these assemblies, we designed two different sets of primers (a total of eight primers) using the NEBilder tool. We extended our three gene fragments with appropriate primers to produce overlap sequences at both ends of each fragment.
To make the improved part, we designed the overlap sequences in such a way that the p2 promoter along with the GFP reporter is flipped. This will make the direction of pBAD promoter and P2 promoter opposite to each other, which will significantly decrease the leaky gene expression of the GFP reporter.
-----Charaterisation-----
Introduction
We observed that there is leaky gene expression in the Unidirectional circuit. We modified the existing part to reduce the leaky gene expression. By flipping the gene we expect that the leaky gene expression will be reduced. To check our hypothesis, we have compared the expression of GFP in the unidirectional and the bidirectional genetic circuit.
Experiment
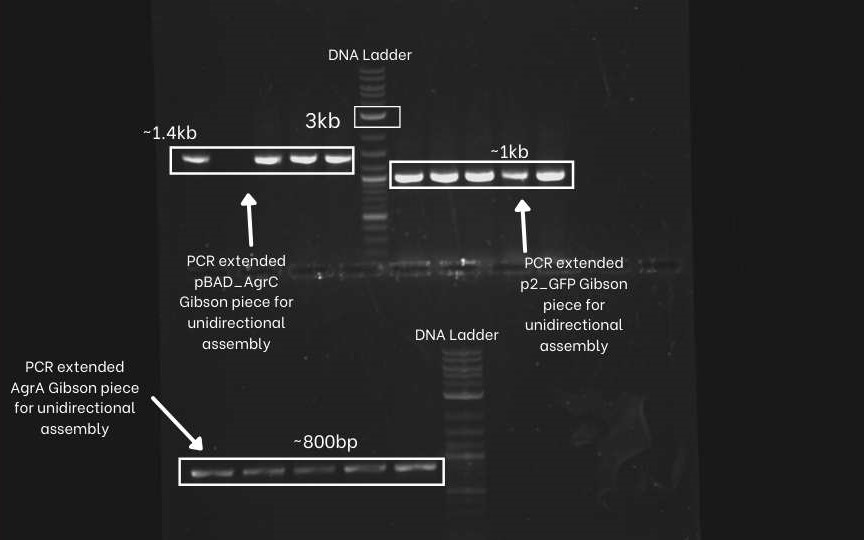
First, we PCR extended the three DNA segments (pBAD-agrC, P2-GFP,agrA,) of the unidirectional circuit with specific primers. To confirm the success of PCR extension an Agarose Gel Electrophoresis was performed. As evident from the picture below, correct bands for all three amplified DNA segments were obtained. (Extended pBAD-agrC ~ 1.4 Kb, Extended P2-GFP ~ 1Kb, Extended agrA ~ 800bp and extended backbone~2kb.
After that, the same four segments (pBAD-agrC, P2-GFP, agrA, and psb1C3 plasmid backbone) were PCR amplified with different sets of specific primers. This extended overlap helps flip the P2-GFP fragments. From the gel electrophoresis image below, we can observe similar band sizes as above because the difference in length of the extension is very low.
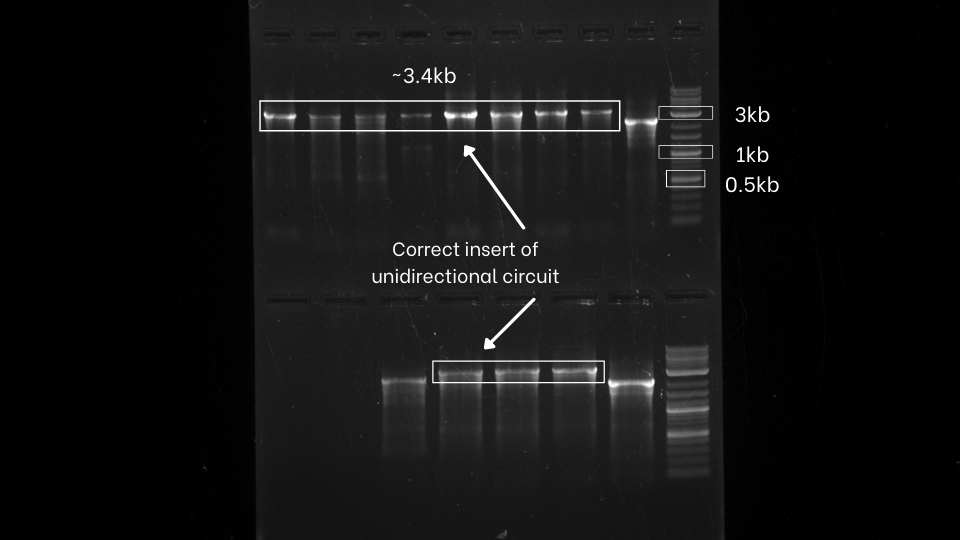
After the successful extension of all the segments, those extended fragments were assembled using Hi-Fi DNA assembly master mix. The reaction mixture was transformed in the E.Coil DH5a. After that, colony PCR of both the unidirectional and bidirectional circuit was performed using VF2 and VR primers to identify the bacterial colony with our desired gene circuits.
As evident from the colony PCR results, positive inserts for both unidirectional and bidirectional assemblies were obtained.
After the successful assembling of both the fragments, we planned to see the expression level of GFP in both bidirectional and unidirectional circuits. For that, first the AIP-1 generator (BBa_K3799006) was expressed in 40ml of bacterial culture to obtain AIP-1 molecules. After expressing for 8-10 hours, the culture was centrifuged and the supernatant was filter-sterilized. The supernatant was lyophilized to concentrate the AIP-1 molecule. After full lyophilization, it was resuspended in 3 ml of NFW to make a stock solution of AIP1.
The stock solution of AIP-1 molecules was diluted to make solutions of different concentrations, from 0% to 100%, where 0% was the negative control and 100% was the one-third dilution of our stock solution.
The two types of cells were incubated (one with unidirectional plasmid and the other with bidirectional plasmid) at 30℃ for 8 hours without AIP-1.
The fluorescence intensity of those two types of cells in absence of AIP-1 molecules was measured at different time points and it was observed that there is a higher fluorescence intensity in cells with unidirectional circuits as compared to the cells with bidirectional circuits. This infers that the bidirectional circuit has less leaky gene expression than that of the unidirectional circuit.
We also wanted to check whether this difference in leaky gene expression is consistent with different concentrations of AIP-1.
To check this, we induced our two types of cells (one with unidirectional plasmid and the other with bidirectional plasmid) with different concentrations of the AIP-1 solution that we mentioned above. From our data, we infer that this leaky gene expression of the unidirectional circuit is consistent in different concentrations of AIP-1 molecules. Also, we can observe a slight decrease in the gap of expression of GFP as the concentration of AIP-1 increases. This may be because of the fact that the ratio of leaky gene expression and the AIP-1 induced expression decreases as the AIP-1 concentration increases.
Final Comments:
Thus we can conclude that our new part BBa_K3799057 is indeed an improvement of the existing part BBa_K1022100 in significantly decreasing the leaky gene expression under the P2 promoter