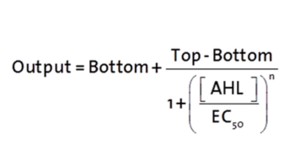Difference between revisions of "Part:BBa K1216007"
(→Characterization of the pLuxR variant dose response to OHHL using plate reader analysis and FACS single cell analysis) |
m (deleted typo (usage and biology heading) |
||
| (51 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1216007 short</partinfo> | <partinfo>BBa_K1216007 short</partinfo> | ||
| − | BBa_K1216007 is a variant of the wild-type | + | BBa_K1216007 is a variant of the wild-type luxR promoter with a lowered sensitivity for LuxR-AHL. This promoter served as a starting point for rational design of a [https://parts.igem.org/Part:BBa_K1216008 library] of eight P<sub>LuxR</sub> variants with differing EC<sub>50</sub> values. (All library member registry entries are linked here: [https://parts.igem.org/Part:BBa_K1216007 1] [https://parts.igem.org/Part:BBa_K1216008 2] [https://parts.igem.org/Part:BBa_K1216009 3] [https://parts.igem.org/Part:BBa_K1216010 4] [https://parts.igem.org/Part:BBa_K1216011 5] [https://parts.igem.org/Part:BBa_K1216012 6] [https://parts.igem.org/Part:BBa_K1216013 7] [https://parts.igem.org/Part:BBa_K1216014 8].) |
| − | [[File:wt_g1_comparison.png|thumb| Comparison of BBa_R0062 to the mutant]] | + | [[File:wt_g1_comparison.png|left|500px|thumb|<b>Figure 1. Comparison of BBa_R0062 to the mutant</b>]] |
| + | <br clear="all"/> | ||
| + | <!-- Add more about the biology of this part here--> | ||
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | [[File:lux_system_ethz.png|thumb|350px| <b>Figure 2: Overview of the Lux signaling system.</b> With no LuxR-AHL complex bound, P<sub>LuxL</sub> enables transcription of <i>luxR</i> whereas P<sub>LuxR</sub> is shut off except for slight expression of the downstream operon. If AHL concentration rises inside the cell, P<sub>LuxL</sub> is shut down and P<sub>LuxR</sub> induced, yielding high expression of LuxI and thus further increase in AHL concentration, constituting positive feedback. *''Placeholder downstream gene.'']] | ||
| + | The Lux signaling system is based on positive feedback. The P<sub>LuxL</sub> promoter drives expression of the upstream <i>luxR</i> gene. The promoter P<sub>LuxR</sub> driver expression of the downstream operon including the LuxI AHL synthetase. The promoter encompasses a palindromic operator sequence to which a dimer of LuxR-AHL complexes binds to induce transcription. In absence of this complex, P<sub>LuxL</sub> is active and thus LuxR is abundant. P<sub>LuxR</sub> however is inactive, showing slight leakage, yielding a fairly low expression level of LuxI and other downstream gene products. | ||
| + | |||
| + | An important feature of AHL, the "signaling molecule" of the system, is that it diffuses freely through cell walls and surrounding media. This enables communication between cells in vincinity. When a certain cell concentration is reached, the diffusing AHL yields sufficiently high levels of the LuxR-AHL complex within cells. The direct consequence of this is high expression of downstream genes, importantly inculding LuxI (and not so importantly shutting down P<sub>LuxL</sub>). Now the positive feedback loop is being closed by elevated synthesis of AHL which further increases LuxI synthesis in cells. | ||
| + | |||
| + | In V. Fischeri other downstream genes include a luciferase which is part of a symbiosis with marine animals. Other possibilities are proteins that facilitate biofilm formation and related phenomena. | ||
| + | |||
| + | In (synthetic) biology, this system can be used as a sender/reporter system by separating LuxI and LuxR, where LuxI is not under control of P<sub>LuxR</sub>, rendering the system independent of cell concentration. LuxR/P<sub>LuxR</sub> can then be used to drive expression of a myriad of other genes such as various reporter genes. | ||
| + | |||
| + | <!-- --> | ||
| + | <span class='h3bb'>Sequence and Features</span> | ||
| + | <partinfo>BBa_K1216008 SequenceAndFeatures</partinfo> | ||
<!-- --> | <!-- --> | ||
| Line 17: | Line 30: | ||
<partinfo>BBa_K1216007 parameters</partinfo> | <partinfo>BBa_K1216007 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | ==Characterization== | ||
| − | + | <b>The final construct was sequenced.</b> | |
| − | [[File:Promoterscheme.jpg|700px|left|thumb|<b>Figure | + | === Site saturation directed mutagenesis of the BBa_R0062 pluxR wild type promoter to obtain the pluxR variant BBa_K2126007=== |
| + | |||
| + | [[File:Promoterscheme.jpg|700px|left|thumb|<b>Figure 2.Location of the site directed saturation mutagenesis in the ''lux''box. </b>As indicated you can see the mutations of the pluxR variant compared to the wild type in red. The positions which were mutated randomly are indicated in red-underlined]] | ||
<br clear="all"/> | <br clear="all"/> | ||
Our goal was to achieve mutations in the luxbox of the pLuxR BBa_R0062 promoter to shift the dose response curve of the promoter depending on the OHHL (3-oxo-hexanoyl-l-homoserine-l-lactone) concentration. | Our goal was to achieve mutations in the luxbox of the pLuxR BBa_R0062 promoter to shift the dose response curve of the promoter depending on the OHHL (3-oxo-hexanoyl-l-homoserine-l-lactone) concentration. | ||
| − | <br>We did site directed mutagenisis of specific sites of the | + | <br>We did site directed saturation mutagenisis of specific sites of the ''lux''box according to literature ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2446796/ Luis Caetano A Mutational Analysis Defines Vibrio fischeri LuxR Binding Sites])<br>We mutated the promoter directly in the BBa_J09855 construct and cloned a eGFP gene as reporter to be able to screen for different dose response curves. In the end we isolate one mutated promoter which shows a shift in sensitivity. For more details see: <i>Characterization of the pLuxR variant dose response to OHHL using plate reader analysis and single cell analysis</i> |
<br clear="all"/> | <br clear="all"/> | ||
| − | ===Characterization of the | + | ===Characterization of the pluxR variant dose response to OHHL using plate reader analysis and single cell analysis=== |
| − | + | <b>Fluorescence data analysis</b> | |
| − | <b>Characterization and comparison to the wild type | + | [[File:300-Formulapluxr1.png|left|300px]] |
| − | [[File: | + | <p>The fitting of the following graphs was performed using this equation :<br><br> |
| − | The | + | Output = eGFP levels [au]<br> |
| + | Top = maximal eGFP level [au]("full induction")<br> | ||
| + | Bottom = minimal eGFP level [au](“leakiness”)<br> | ||
| + | n = Hill coefficient (“cooperativity”)<br> | ||
| + | EC<sub>50</sub> = Half-maximal effective concentration (“sensitivity”)<br> | ||
| + | [AHL]=OHHL concentration [nM]</p> | ||
| + | <br clear="all"/> | ||
| + | <b>Characterization and comparison to the wild type pluxR using microtiter plate analysis</b> | ||
| + | [[File:Wtg1microtiter.jpg|400px|left|thumb|<b>Figure 3. Sensitivity curve of the wild type promoter and pluxR variant (G1)</b> analyzed on the Tecan M200 Infinity plate reader. The OHHL concentration range is plotted against the standarized fluorescence (au).<br>For the wild type : EC<sub>50</sub>=5.86 nM, R<sup>2</sup>= 0.87,n=1.7 <br>Fore The G1 mutant : EC<sub>50</sub>=1'341 nM, R<sup>2</sup>= 0.98,n=0. All assays were carried out in triplicates, results are presented as mean ± standard deviation.]] | ||
| + | The eGFP reporter allows us to make fluorescence measurement in plate reader experiments as well as in single cell analysis to characterize the promoter behavior depending on [OHHL], by recording the fluorescence [au]. According to literature [http://www.ncbi.nlm.nih.gov/pubmed/18760602 (Geske G.D. Evaluation of a focused library of N-aryl L-homoserine lactones reveals a new set of potent quorum sensing modulators)] we could define the first experimental OHHL ranges to induce the promoter. After some fine tunning and adjusting experiment we could narrow down the ranges to obtain a dose response curve in a reasonable resolution for the BBa_R0062 wild type promoter to the OHHL (3-oxo-hexanoyl-l-homoserine-lactone) concentration. See Figure 3. these data were obtained from a plate reader experiment. According to the sensitivity range of the wild type promoter we could screen for promoters, and scan the potentially shifted sensitivity variants in different ranges of [OHHL]. We finally isolated one promoter variant which shows a shift in sensitivity compared to the wild type. See G1 on <i>Figure 1</i>. The pluxR variant has an EC<sub>50</sub>=1'341 nM , the pluxR wild type has an EC<sub>50</sub>=5.86 nM according to the plate reader data. The single cell analysis gave us data of higher quality (see below). | ||
<br clear="all"/> | <br clear="all"/> | ||
| − | <b>Characterization of the | + | <b>Characterization of the pluxR dose response to OHHL in liquid culture and in agar plates by single cell analysis</b><br><br> |
| − | [[File:facsdatag1.jpg|700px| | + | <p>To obtain high quality data we did single cell analysis over the range defined above. As expected the EC<sub>50</sub> change compared to the microtiter plate data. <br> See Figure 4. </p> |
| − | + | [[File:facsdatag1.jpg|700px|left|thumb|<b>Figure 4. Dose response comparison between [OHHL] induction in liquid culture and on agar plates</b>, of the BBa_J09855 construct using the pluxR variant BBa_K1216007 promoter and a eGFP gene as reporter</b> analyzed at different [OHHL] in the single cell analysis. The black curve shows the response of to OHHL in liquid culture, the orange curve shows the response to OHHL on agar plates. All assays were carried out in duplicates, results are presented as mean ± standard deviation.]] | |
| − | [[File:AgaLiG1.jpg|400px|left|thumb|<b>Figure | + | <br clear="all"/> |
| + | [[File:AgaLiG1.jpg|400px|left|thumb|<b>Figure 5. Dose response curve of the pluxR variant G1 on agar plates and liquid culture.</b> The black curve shows the response of to OHHL in liquid culture, the orange curve shows the response to OHHL on agar plates<br>For the liquid culture we got :EC<sub>50</sub>=6'482nM, R<sup>2</sup>=0.97, n=0.8;<br>For the agar plates we got :EC<sub>50</sub>=12'555nM, R<sup>2</sup>=0.93, n=0.8. All assays were carried out in duplicates, results are presented as mean ± standard deviation.]] | ||
| + | <p>Figure 5 shows the dose reponse curve of the promoter depending on OHHL concentration. We performed this experiment in liquid culture as well as in agar plates (OHHL was added to the melted 1.5% agar), which was closer to our project. We observed a shift in the dose response between the liquid culture (EC<sub>50</sub>=6'482 nM) single cell analysis and the one in agar plates(EC<sub>50</sub>=12'555 nM).This correspond to a 2 fold shift.</p> | ||
<br clear="all"/> | <br clear="all"/> | ||
| − | <b>Comparison of the | + | <b>Comparison of the pluxR variant and pluxR wild type dose response to OHHL in liquid culture and on agar plates</b> |
| − | [[File: | + | [[File:facscomparisonplate.jpg|400px|left|thumb|<b>Figure 6. Comparison of the sensitivity of the wild type promoter and the mutated PluxR1 promoter on agar plates</b><br>For the wild type we got :EC<sub>50</sub>=4.45nM, R<sup>2</sup>=0.80, n=1.7<br>For the pluxR variant we got: EC<sub>50</sub>=12'555 nM, R<sup>2</sup>=0.93, n=0.8. |
| + | All assays were carried out in duplicates, results are presented as mean ± standard deviation.]] | ||
| + | [[File:facscomparisonliquid.jpg|400px|right|thumb|<b>Figure 7. Comparison of the sensitivity of the wild type promoter and the mutated PluxR1 promoter in liquid culture</b><br>For the wild type we got: EC<sub>50</sub>=0.02 nM, R<sup>2</sup>=0.84, n=1.7<br>For the pLuxR variant we got: EC<sub>50</sub>=6'462 nM, R<sup>2</sup>=0.97, n=0.8. All assays were carried out in duplicates, results are presented as mean ± standard deviation.]] | ||
<br clear="all"/> | <br clear="all"/> | ||
| − | + | <p>Figure 6 shows the comparison between the wild type pluxR and pluxR variant dose reponse curve to [OHHL] on agar plates (OHHL was added to the melted 1.5% agar), which was closer to our project. The shift in the dose response between the wild type pluxR (EC<sub>50</sub>=4.45 nM and the pluxR variant(EC<sub>50</sub>=12'555 nM) is about 2800 times less sensitive according to the single cell analysis. See <i>Figure 4</i></p><br><br> | |
| + | <p>Figure 7. shows the comparison between the wild type pluxR and pluxR variant dose reponse curve to [OHHL] in liquid cultures. The shift in the dose response between the wild type pluxR (EC<sub>50</sub>=0.02 nM and the pluxR variant(EC<sub>50</sub>=6'462 nM) is about 300'000 times different according to the single cell analysis. See <i>Figure 4</i></p> | ||
| + | <br clear="all"/> | ||
| + | |||
| + | ===References=== | ||
| + | (1) M Geske G.D.,Evaluation of a focused library of N-aryl L-homoserine lactones reveals a new set of potent quorum sensing modulators. | ||
| + | (2) Luis Caetano A Mutational Analysis Defines Vibrio fischeri LuxR Binding Sites | ||
Latest revision as of 20:33, 10 June 2014
Variant of the wild-type pLuxR promoter with lower sensitivity
BBa_K1216007 is a variant of the wild-type luxR promoter with a lowered sensitivity for LuxR-AHL. This promoter served as a starting point for rational design of a library of eight PLuxR variants with differing EC50 values. (All library member registry entries are linked here: 1 2 3 4 5 6 7 8.)
Usage and Biology

The Lux signaling system is based on positive feedback. The PLuxL promoter drives expression of the upstream luxR gene. The promoter PLuxR driver expression of the downstream operon including the LuxI AHL synthetase. The promoter encompasses a palindromic operator sequence to which a dimer of LuxR-AHL complexes binds to induce transcription. In absence of this complex, PLuxL is active and thus LuxR is abundant. PLuxR however is inactive, showing slight leakage, yielding a fairly low expression level of LuxI and other downstream gene products.
An important feature of AHL, the "signaling molecule" of the system, is that it diffuses freely through cell walls and surrounding media. This enables communication between cells in vincinity. When a certain cell concentration is reached, the diffusing AHL yields sufficiently high levels of the LuxR-AHL complex within cells. The direct consequence of this is high expression of downstream genes, importantly inculding LuxI (and not so importantly shutting down PLuxL). Now the positive feedback loop is being closed by elevated synthesis of AHL which further increases LuxI synthesis in cells.
In V. Fischeri other downstream genes include a luciferase which is part of a symbiosis with marine animals. Other possibilities are proteins that facilitate biofilm formation and related phenomena.
In (synthetic) biology, this system can be used as a sender/reporter system by separating LuxI and LuxR, where LuxI is not under control of PLuxR, rendering the system independent of cell concentration. LuxR/PLuxR can then be used to drive expression of a myriad of other genes such as various reporter genes.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
The final construct was sequenced.
Site saturation directed mutagenesis of the BBa_R0062 pluxR wild type promoter to obtain the pluxR variant BBa_K2126007
Our goal was to achieve mutations in the luxbox of the pLuxR BBa_R0062 promoter to shift the dose response curve of the promoter depending on the OHHL (3-oxo-hexanoyl-l-homoserine-l-lactone) concentration.
We did site directed saturation mutagenisis of specific sites of the luxbox according to literature ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2446796/ Luis Caetano A Mutational Analysis Defines Vibrio fischeri LuxR Binding Sites])
We mutated the promoter directly in the BBa_J09855 construct and cloned a eGFP gene as reporter to be able to screen for different dose response curves. In the end we isolate one mutated promoter which shows a shift in sensitivity. For more details see: Characterization of the pLuxR variant dose response to OHHL using plate reader analysis and single cell analysis
Characterization of the pluxR variant dose response to OHHL using plate reader analysis and single cell analysis
Fluorescence data analysis
The fitting of the following graphs was performed using this equation :
Output = eGFP levels [au]
Top = maximal eGFP level [au]("full induction")
Bottom = minimal eGFP level [au](“leakiness”)
n = Hill coefficient (“cooperativity”)
EC50 = Half-maximal effective concentration (“sensitivity”)
[AHL]=OHHL concentration [nM]
Characterization and comparison to the wild type pluxR using microtiter plate analysis

For the wild type : EC50=5.86 nM, R2= 0.87,n=1.7
Fore The G1 mutant : EC50=1'341 nM, R2= 0.98,n=0. All assays were carried out in triplicates, results are presented as mean ± standard deviation.
The eGFP reporter allows us to make fluorescence measurement in plate reader experiments as well as in single cell analysis to characterize the promoter behavior depending on [OHHL], by recording the fluorescence [au]. According to literature [http://www.ncbi.nlm.nih.gov/pubmed/18760602 (Geske G.D. Evaluation of a focused library of N-aryl L-homoserine lactones reveals a new set of potent quorum sensing modulators)] we could define the first experimental OHHL ranges to induce the promoter. After some fine tunning and adjusting experiment we could narrow down the ranges to obtain a dose response curve in a reasonable resolution for the BBa_R0062 wild type promoter to the OHHL (3-oxo-hexanoyl-l-homoserine-lactone) concentration. See Figure 3. these data were obtained from a plate reader experiment. According to the sensitivity range of the wild type promoter we could screen for promoters, and scan the potentially shifted sensitivity variants in different ranges of [OHHL]. We finally isolated one promoter variant which shows a shift in sensitivity compared to the wild type. See G1 on Figure 1. The pluxR variant has an EC50=1'341 nM , the pluxR wild type has an EC50=5.86 nM according to the plate reader data. The single cell analysis gave us data of higher quality (see below).
Characterization of the pluxR dose response to OHHL in liquid culture and in agar plates by single cell analysis
To obtain high quality data we did single cell analysis over the range defined above. As expected the EC50 change compared to the microtiter plate data.
See Figure 4.


For the liquid culture we got :EC50=6'482nM, R2=0.97, n=0.8;
For the agar plates we got :EC50=12'555nM, R2=0.93, n=0.8. All assays were carried out in duplicates, results are presented as mean ± standard deviation.
Figure 5 shows the dose reponse curve of the promoter depending on OHHL concentration. We performed this experiment in liquid culture as well as in agar plates (OHHL was added to the melted 1.5% agar), which was closer to our project. We observed a shift in the dose response between the liquid culture (EC50=6'482 nM) single cell analysis and the one in agar plates(EC50=12'555 nM).This correspond to a 2 fold shift.
Comparison of the pluxR variant and pluxR wild type dose response to OHHL in liquid culture and on agar plates

For the wild type we got :EC50=4.45nM, R2=0.80, n=1.7
For the pluxR variant we got: EC50=12'555 nM, R2=0.93, n=0.8. All assays were carried out in duplicates, results are presented as mean ± standard deviation.

For the wild type we got: EC50=0.02 nM, R2=0.84, n=1.7
For the pLuxR variant we got: EC50=6'462 nM, R2=0.97, n=0.8. All assays were carried out in duplicates, results are presented as mean ± standard deviation.
Figure 6 shows the comparison between the wild type pluxR and pluxR variant dose reponse curve to [OHHL] on agar plates (OHHL was added to the melted 1.5% agar), which was closer to our project. The shift in the dose response between the wild type pluxR (EC50=4.45 nM and the pluxR variant(EC50=12'555 nM) is about 2800 times less sensitive according to the single cell analysis. See Figure 4
Figure 7. shows the comparison between the wild type pluxR and pluxR variant dose reponse curve to [OHHL] in liquid cultures. The shift in the dose response between the wild type pluxR (EC50=0.02 nM and the pluxR variant(EC50=6'462 nM) is about 300'000 times different according to the single cell analysis. See Figure 4
References
(1) M Geske G.D.,Evaluation of a focused library of N-aryl L-homoserine lactones reveals a new set of potent quorum sensing modulators. (2) Luis Caetano A Mutational Analysis Defines Vibrio fischeri LuxR Binding Sites