Difference between revisions of "Part:BBa K818000:Experience"
(→User Reviews) |
|||
| (16 intermediate revisions by the same user not shown) | |||
| Line 12: | Line 12: | ||
|- | |- | ||
|width='10%'| | |width='10%'| | ||
| − | <partinfo> | + | <partinfo>BBa_K818000 AddReview number</partinfo> |
<I>UsernEnter the review inofrmation here.ame</I> | <I>UsernEnter the review inofrmation here.ame</I> | ||
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
| Line 22: | Line 22: | ||
|- | |- | ||
|width='10%'| | |width='10%'| | ||
| − | <partinfo> | + | <partinfo>BBa_K818000 1</partinfo> |
<I>Newcastle University iGEM 2013</I> | <I>Newcastle University iGEM 2013</I> | ||
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
| Line 52: | Line 52: | ||
<br> | <br> | ||
<table border="0"> | <table border="0"> | ||
| − | <td><img src="https://static.igem.org/mediawiki/parts/1/10/BareCillus_Rrnb_1.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">Plate 1: B. Subtilis str. 168 transformed with | + | <td><img src="https://static.igem.org/mediawiki/parts/1/10/BareCillus_Rrnb_1.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">Plate 1: B. Subtilis str. 168 transformed with pGFPrrnB.</div></td> |
| − | <td><img src="https://static.igem.org/mediawiki/parts/8/89/BareCillus_H20_1.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">Plate 2: B.subtilis str. 168 transformed with | + | <td><img src="https://static.igem.org/mediawiki/parts/8/89/BareCillus_H20_1.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">Plate 2: B.subtilis str. 168 transformed with H20(negative control).</div></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td><img src="https://static.igem.org/mediawiki/parts/6/67/BareCillus_Gro_1.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">Plate 3: B. subtilis str. 168 | + | <td><img src="https://static.igem.org/mediawiki/parts/6/67/BareCillus_Gro_1.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">Plate 3: B. subtilis str. 168 with pSac + Cm derived plasmid. |
</div></td> | </div></td> | ||
| − | <td><div class="italic">Figure 1. Plates of B. Subtilis str. 168 transformed with | + | <td><div class="italic">Figure 1. Plates of B. Subtilis str. 168 transformed with the pSac+Cm derived plasmid, pGFPrrnB (positive control) and water (negative control).</div></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 84: | Line 84: | ||
<body> | <body> | ||
<table border="0"> | <table border="0"> | ||
| − | <td><img src="https://static.igem.org/mediawiki/parts/6/69/BareCillus_Gro5.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic"> | + | <td><img src="https://static.igem.org/mediawiki/parts/6/69/BareCillus_Gro5.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">B. subtilis str. 168 with 5ug pSac + Cm derived plasmid.</div></td> |
| − | <td><img src="https://static.igem.org/mediawiki/parts/7/72/BareCillus_Gro10.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic"> | + | <td><img src="https://static.igem.org/mediawiki/parts/7/72/BareCillus_Gro10.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">B. subtilis str. 168 with 10ug pSac + Cm derived plasmid.</div></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td><img src="https://static.igem.org/mediawiki/parts/7/74/BareCillus_Gro15.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic"> | + | <td><img src="https://static.igem.org/mediawiki/parts/7/74/BareCillus_Gro15.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">B. subtilis str. 168 with 15ug pSac + Cm derived plasmid.</div></td> |
| − | <td><img src="https://static.igem.org/mediawiki/parts/8/89/BareCillus_H20_1.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic"> | + | <td><img src="https://static.igem.org/mediawiki/parts/8/89/BareCillus_H20_1.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">B.subtilis str. 168 transformed with H20(negative control)</div></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td><img src="https://static.igem.org/mediawiki/parts/6/63/Rrnb_2.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic"> | + | <td><img src="https://static.igem.org/mediawiki/parts/6/63/Rrnb_2.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">B. Subtilis str. 168 transformed with pGFPrrnB.</div></td> |
| − | <td><div class="italic">Figure | + | <td><div class="italic">Figure 1. Plates of B. Subtilis str. 168 transformed with 5,10 and 15ug pSac+Cm derived plasmid, pGFPrrnB (positive control) and water (negative control).</div></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 100: | Line 100: | ||
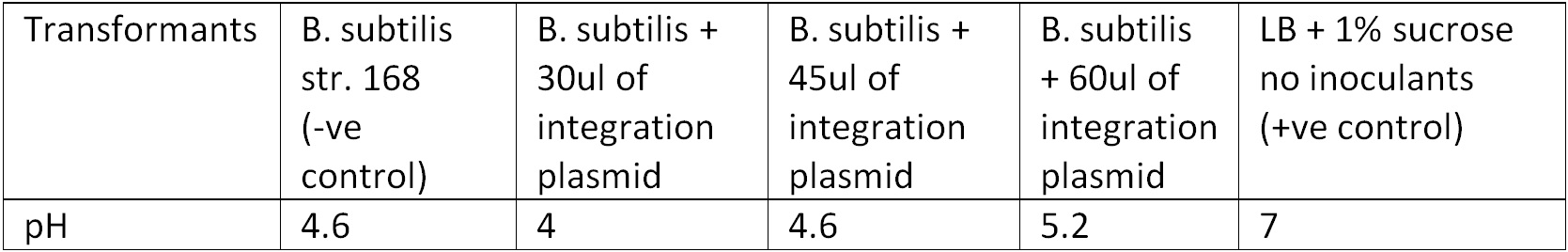
To test for integration, we used the Phenol red Sucrose test; the media where the transformants from the 10ug and 15ug plates were inoculated showed the same results as the control (‘’B. subtilis’’ + pGFPrrnB) with pH ranging between 4.6 – 5.2 suggesting that they could utilized sucrose as their carbon source and produced the acid by products, Figure 3. | To test for integration, we used the Phenol red Sucrose test; the media where the transformants from the 10ug and 15ug plates were inoculated showed the same results as the control (‘’B. subtilis’’ + pGFPrrnB) with pH ranging between 4.6 – 5.2 suggesting that they could utilized sucrose as their carbon source and produced the acid by products, Figure 3. | ||
| − | + | ||
| − | + | [[File:BareCillus_Gro_pH.jpg|900px]] | |
| − | + | ||
| − | + | ||
Figure 3. Display the pH of each samples following the Phenol Red Sucrose test on overnight grown culture in LB + 1% Sucrose media. | Figure 3. Display the pH of each samples following the Phenol Red Sucrose test on overnight grown culture in LB + 1% Sucrose media. | ||
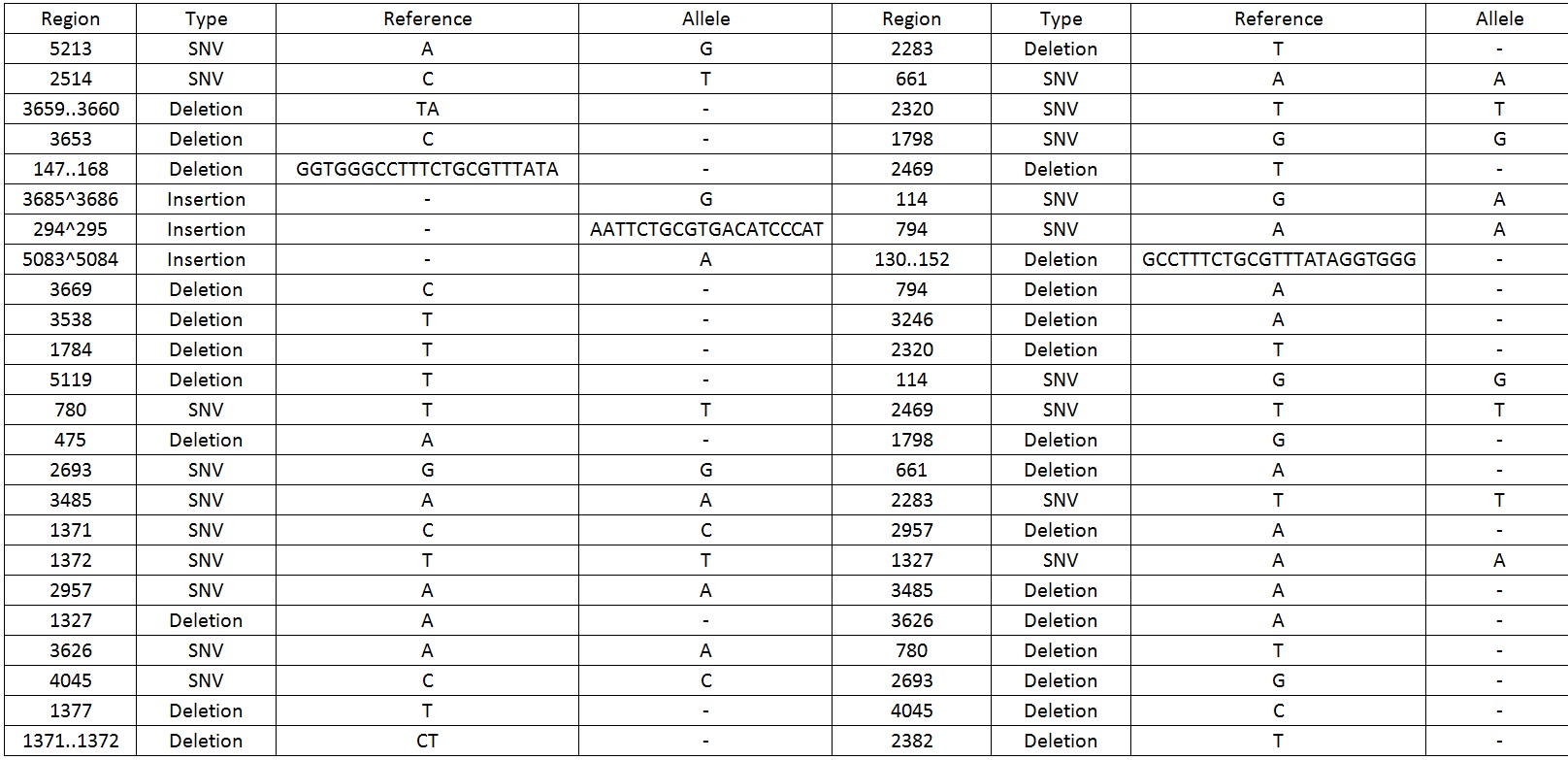
| − | We then sequenced the backbone and found out that there were 49 mutations which include SNV, deletions and insertions | + | We then sequenced the backbone and found out that there were 49 mutations which include SNV, deletions and insertions.Table 1 display the mutations found on the sequence. |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | [[File:BareCillus_Gro_mutations.jpg|700px]] | ||
Table 1. Shows all the mutations found in the BBa_K818000 backbone. | Table 1. Shows all the mutations found in the BBa_K818000 backbone. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
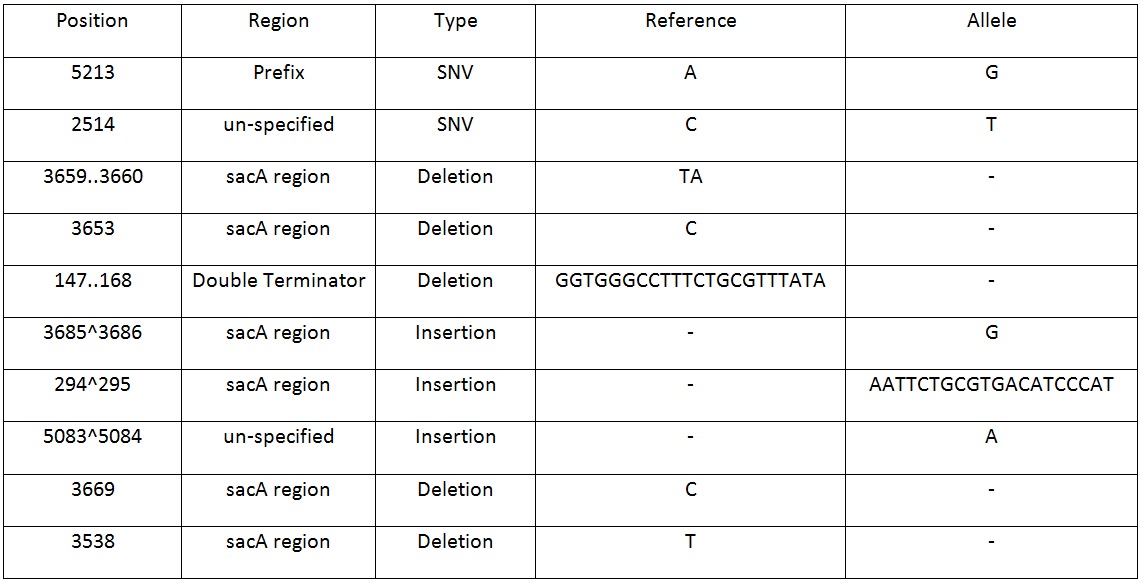
Table 2. Shows list of mutations found in the BBa_K818000 backbone, including the position, region and type of mutations after analysing the initial sequencing and the sequencing of highly mutated regions. | Table 2. Shows list of mutations found in the BBa_K818000 backbone, including the position, region and type of mutations after analysing the initial sequencing and the sequencing of highly mutated regions. | ||
| + | [[File:BareCillus Gro sumseq.jpg|700px]] | ||
| + | Table 2. Shows list of mutations found in the BBa_K818000 backbone, including the position, region and type of mutations after analysing the initial sequencing and the sequencing of highly mutated regions. | ||
| + | |||
| + | [[File:Gro analysis high thgrougput.png|700px]] | ||
| + | |||
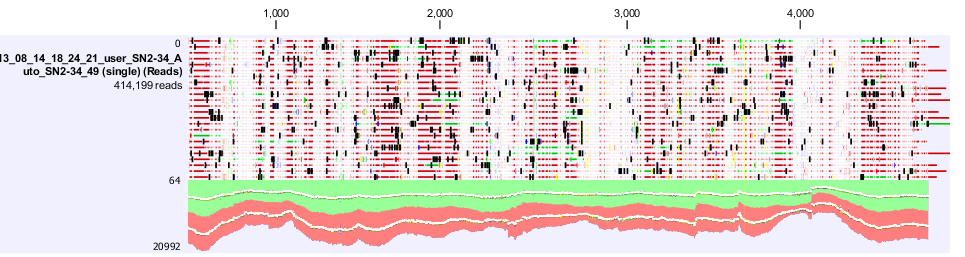
| + | Figure 4. shows a screen shot of the pSac-Cm derived integration plasmid sequence analysis that we performed. It shows the amount of coverage, where the mutations are. | ||
The results from both sequencing run proved to show similar mutations were found on this backbone, most of the mutations occurred in the sacA integration regions. These results explained the reason why this pSac-Cm derived integration backbone for ‘’B.subtilis’’ were not working. | The results from both sequencing run proved to show similar mutations were found on this backbone, most of the mutations occurred in the sacA integration regions. These results explained the reason why this pSac-Cm derived integration backbone for ‘’B.subtilis’’ were not working. | ||
Latest revision as of 17:45, 4 October 2013
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K818000
This backbone plasmid was used as primary backbone for all constructs in team Groningen 2012 project: the Food Warden. For further info: [http://2012.igem.org/Team:Groningen/OurBiobrick iGEM Groningen 2012 biobrick page]
User Reviews
UNIQ2b41959173419081-partinfo-00000000-QINU