Difference between revisions of "Part:BBa K1100000"
(→Quantitative Measurement Data) |
(→Quantitative Measurement Data) |
||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 37: | Line 37: | ||
[[File:Aleader3.jpg|400px|thumb|center|'''Figure 3'''. Mutation positions of Mut-ALeader [B-F] switch plot of some mutants]] | [[File:Aleader3.jpg|400px|thumb|center|'''Figure 3'''. Mutation positions of Mut-ALeader [B-F] switch plot of some mutants]] | ||
| − | With fluorescent measurement data, we | + | With fluorescent measurement data, we can conclude that directed mutagenesis is not a effective way to optimize our Aleader part. |
| − | [[File:Aleader4A.jpg|400px|thumb|left|'''Figure 4A.''' Switch Plot of Mut3]] [[File:Aleader4B.jpg|400px|thumb|right|'''Figure 4B'''Switch Plot of Mut8]] | + | So we further support our optimization of our Aleader with a triphase model(To learn more about that, click HERE). |
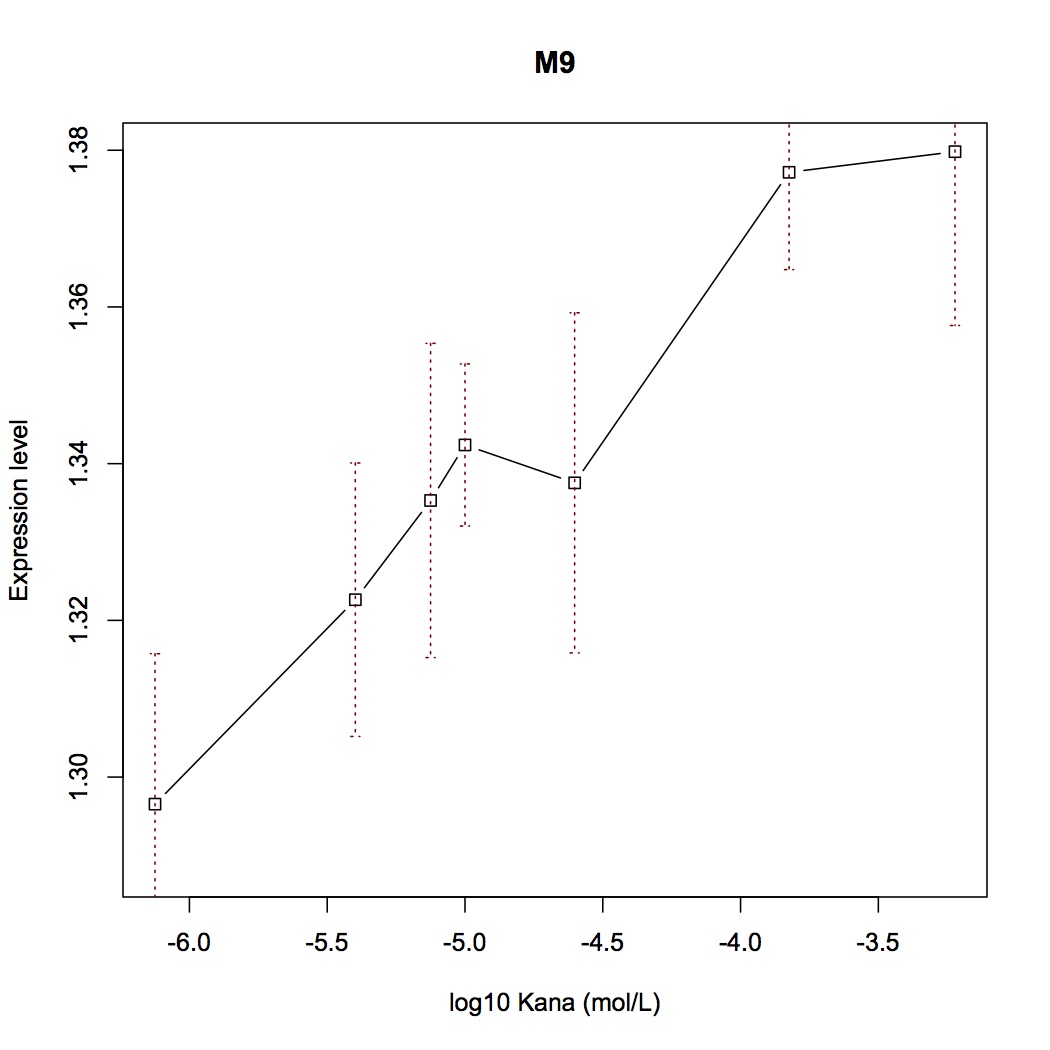
| + | [[File:Aleader4A.jpg|400px|thumb|left|'''Figure 4A.''' Switch Plot of Mut3]] [[File:Aleader4B.jpg|400px|thumb|right|'''Figure 4B.'''Switch Plot of Mut8]] | ||
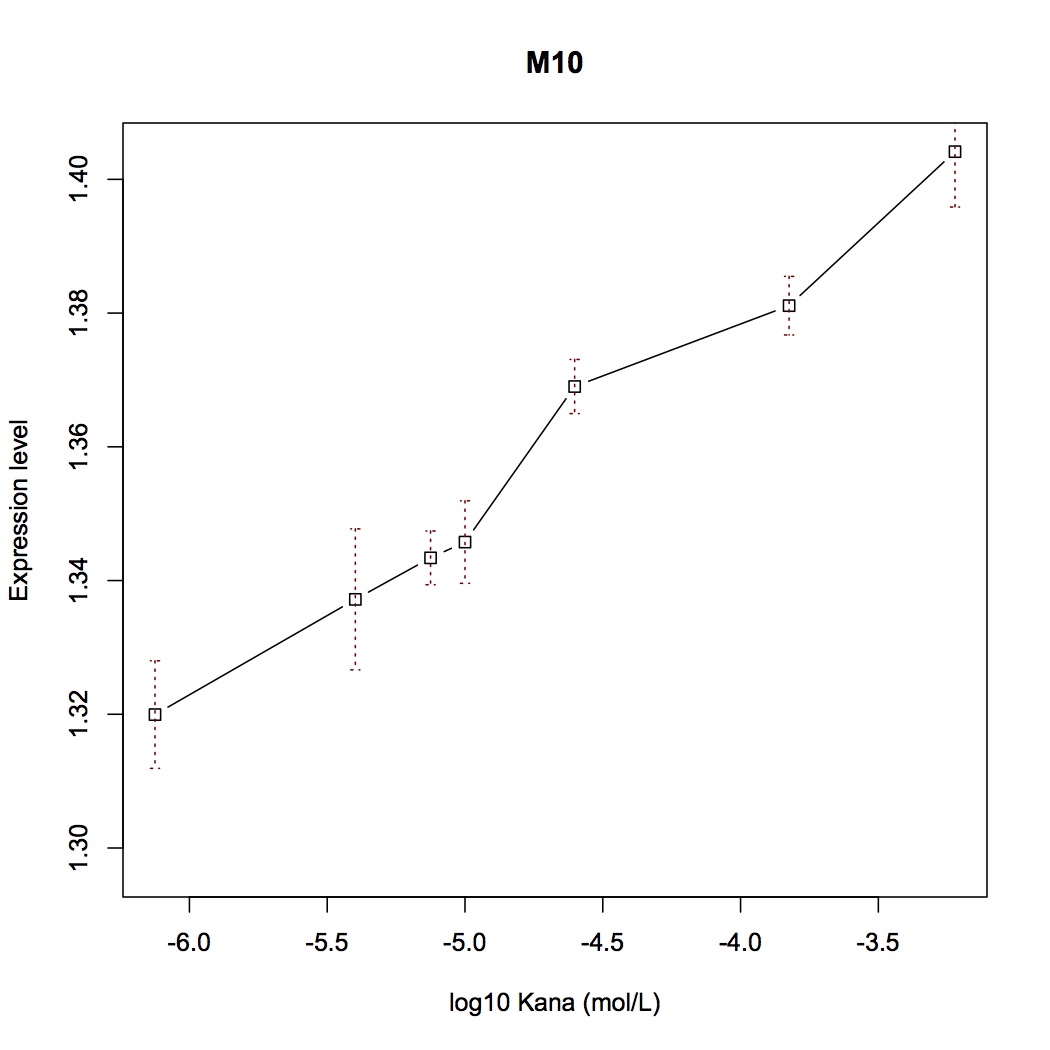
| − | [[File:Aleader4C.jpg|400px|thumb|left|'''Figure 4C.''' Switch Plot of Mut9]] [[File:Aleader4D.jpg|400px|thumb|right|'''Figure 4D'''Switch Plot of Mut10]] | + | [[File:Aleader4C.jpg|400px|thumb|left|'''Figure 4C.''' Switch Plot of Mut9]] [[File:Aleader4D.jpg|400px|thumb|right|'''Figure 4D.'''Switch Plot of Mut10]] |
[[File:Aleader4E.jpg|400px|thumb|left|'''Figure 4E.''' Switch Plot of Mut11]] | [[File:Aleader4E.jpg|400px|thumb|left|'''Figure 4E.''' Switch Plot of Mut11]] | ||
| + | |||
| + | == Reference == | ||
| + | [1] Jia X, Zhang J, Sun W, et al. Riboswitch Control of Aminoglycoside Antibiotic Resistance[J]. Cell, 2013, 152(1): 68-81. | ||
| + | |||
| + | [2] Hanau‐Berçot B, Podglajen I, Casin I, et al. An intrinsic control element for translational initiation in class 1 integrons[J]. Molecular microbiology, 2002, 44(1): 119-130. | ||
Latest revision as of 01:51, 28 September 2013
ALeader
A kind of regulatory part that is induced by aminoglycoside drug.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Background
Riboswitches are regulatory RNAs that regulate gene expression by binding small molecule metabolites. Recently, a novel riboswitch was reported which is present in the leader RNA of the aminoglycoside resistance genes that encode the aminoglycoside acetyl transferase (AAC) and aminoglycoside adenyl transferase (AAD) enzymes. When the aminoglycosides bind to the leader RNA, a conformational change will be induced, leading to the expression of the aminoglycoside resistance genes(Figure 1). (Jia X et al, 2013.) In addition, the leader RNA encompass an integrase site (attl1)which is overlapped by a short peptide encoded by an ORF (ORF11). ( Roth A et al.) Therefore, Aleader is a complicated riboswitch contains ORF11, cistron, aminoglycoside-binding domain and att1 recombination site. Due to the complicacy of Aleader, the secondary structure of it could not be predicted by most software such as Mfold and NUPACK. (Jia X et al, 2013.).
The 75nt RNA sequence of ALeader contains two SD sequences (ribosome binding sites) and an anti-SD sequence (CUUC) which can complementarily pair with either of the SD sequences. In the absence of aminoglycosides, anti-SD pairs with SD2. The binding of ribosomes to SD1 triggers the translation of a small peptide which stops at the stop codon ahead of SD2, therefore inhibits the translation of the gene after SD2. When aminoglycosides (kanamycin for example) exists, it will induce a structural change of Aleader. The anti-SD sequence pairs with SD1, consequently unmasking SD2 for ribosomal binding, which results in the translation of the following gene.
Our Aleader riboswitch demonstrates progressive induction of reporter genes in response to sublethal doses of the antibiotics. It makes sense because an antibiotic-resistant riboswitch must be able to detect low levels of antibiotics and activate the resistance mechanism before the cells are killed. Thus, Aleader turns out to be a novel translation regulatory part with high dynamic range, slight response delay and immense modification potential.
Mutation Design
To improve the natural bio-brick part’s performance, we attempted to modify the sequence of ALeader and adjusted its structure. This library of mutations, hence, is a collection of brand new inducible translational elements.
In vitro experiments revealed that the structural importance of leader RNA lays on three domains: the SD1, SD2 and the anti-SD sequence (Figure 2). Mutations of these domains might change the relative stability of two conformation, thereby increasing or decreasing K1. Meanwhile, the mutations of other sites which might have direct interactions with antibiotic moleculars would alter the ligand binding affinity, leading to the change of K2.

Quantitative Measurement Data
With mutations on these critical sites, the mutual binding affinities and local structual stabilities change thus generating new riboswitch. We selected several point-directed mutations as examples(Figure 3).
With fluorescent measurement data, we can conclude that directed mutagenesis is not a effective way to optimize our Aleader part. So we further support our optimization of our Aleader with a triphase model(To learn more about that, click HERE).
Reference
[1] Jia X, Zhang J, Sun W, et al. Riboswitch Control of Aminoglycoside Antibiotic Resistance[J]. Cell, 2013, 152(1): 68-81.
[2] Hanau‐Berçot B, Podglajen I, Casin I, et al. An intrinsic control element for translational initiation in class 1 integrons[J]. Molecular microbiology, 2002, 44(1): 119-130.