Difference between revisions of "Part:BBa K1100141"
(→Manipulating and Experimental Data (Quantitative Measurement)) |
|||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1100141 short</partinfo> | <partinfo>BBa_K1100141 short</partinfo> | ||
| − | <p></p> | + | <p></p> |
| − | + | ||
| + | <!-- Add more about the biology of this part here | ||
| + | ===Usage and Biology=== | ||
| + | |||
| + | <!-- --> | ||
| + | <span class='h3bb'>Sequence and Features</span> | ||
| + | <partinfo>BBa_K1100141 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | == Background == | ||
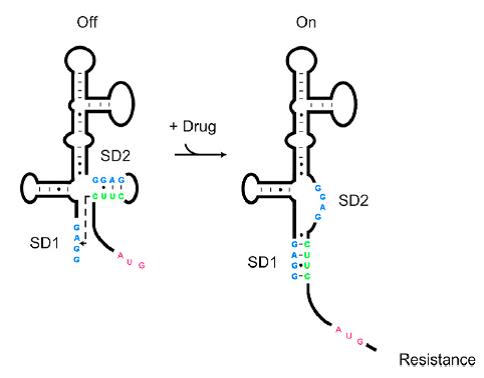
| + | Riboswitches are regulatory RNAs that regulate gene expression by binding small molecules. Recently, a novel riboswitch was reported which is present in the leader RNA of the aminoglycoside resistance genes that encode the aminoglycoside acetyl transferase (AAC) and aminoglycoside adenyl transferase (AAD) enzymes. When the aminoglycosides bind to the leader RNA, a conformational change will be induced, leading to the expression of the aminoglycoside resistance genes (Jia X et al, 2013.). In addition, the leader RNA encompass an integrase site (attl1), which is overlapped by a short peptide encoded by an ORF (ORF11) (Berçot B, et al, 2002.). Therefore, Aleader is a complicated riboswitch containing ORF11, cistron, aminoglycoside-binding domain and att1 recombination site. Due to the complicacy of Aleader, the secondary structure of it could not be predicted by most software such as Mfold and NUPACK (Jia X et al, 2013.). | ||
| + | |||
| + | [[File:AleaderT1.jpg|600px|thumb|center| '''Figure1'''. Proposed Model for the Induction of Aminoglycoside Resistance: Aminoglycoside binding to the leader RNA induces a change in the leader RNA structure(Jia X et al, 2013.).]] | ||
| + | As shown in '''Figure 1''', the 75nt RNA sequence of Aleader contains two SD sequences (ribosome binding sites) and an anti-SD sequence (CUUC) which can complementarily pair with either of the SD sequences. In the absence of aminoglycosides, anti-SD pairs with SD2. The binding of ribosomes to SD1 triggers the translation of a small peptide which stops at the stop codon ahead of SD2, and therefore inhibits the translation of the desired gene after SD2. When aminoglycosides (kanamycin for example) exists, it will induce a structural change of Aleader. The anti-SD sequence base-pairs with SD1, consequently unmasking SD2 for ribosomal binding, which results in the translation of the following gene. | ||
| + | == Our design of AleaderT == | ||
Many riboswitches have lower dynamic range and higher leakage rate compared with protein regulators. To reduce the basal level and enhance the dynamic range, while having no effect on the protein product, we designed a novel riboswitch called ALeaderT, following the principle of three-phase switch design. | Many riboswitches have lower dynamic range and higher leakage rate compared with protein regulators. To reduce the basal level and enhance the dynamic range, while having no effect on the protein product, we designed a novel riboswitch called ALeaderT, following the principle of three-phase switch design. | ||
We added a GGGS linker (dock III), a common neutral peptide linker, after the SD2 and ATG. Impressively, by RNA 2D-prediction, we found that dock III will capture antiSD2 sequence to form a stable stem loop, with characteristics of terminator, though neutral as an amino peptide. | We added a GGGS linker (dock III), a common neutral peptide linker, after the SD2 and ATG. Impressively, by RNA 2D-prediction, we found that dock III will capture antiSD2 sequence to form a stable stem loop, with characteristics of terminator, though neutral as an amino peptide. | ||
| − | We suppose that the ALeaderT will have three different steady-state conformations, A “Translation off Termination off”, B “Translation on Termination off”, and C “Translation on Termination on”('''Figure | + | We suppose that the ALeaderT will have three different steady-state conformations, A “Translation off Termination off”, B “Translation on Termination off”, and C “Translation on Termination on”('''Figure 3'''). |
[[File:AleaderT2.jpg|600px|thumb|center| '''Figure 3'''.Tri-Phase Model of AleaderT.]] | [[File:AleaderT2.jpg|600px|thumb|center| '''Figure 3'''.Tri-Phase Model of AleaderT.]] | ||
| + | The A conformation is the ligand-free conformation that SD2 and antiSD form the stem loop, resulting in no translation nor termination. The B conformation is the ligand-binding conformation that SD2 is released and SD1 binds the antiSD, leading to the translation. And in C conformation, the SD2 is also released, making it possible to bind the ribosome, but the antiSD and dock III form a stable stem loop to reduce the transcription. | ||
| + | Through mathematical modeling, we found that the leakage expression would significantly decrease, while the dynamic range increased several-fold. The working range is usually changed, as well. | ||
| − | + | '''1.Dynamic Range'''(shown with ratio): | |
| − | [[File: | + | [[File:AleaderT3.jpg|1000px|thumb|center| '''Figure 4'''.Triphase riboswitch can break through the upperlimit of a biphase riboswitch when the conformation B is dominant to conformation A. |
| + | ''k2'':distribution constant between different conformations ]] | ||
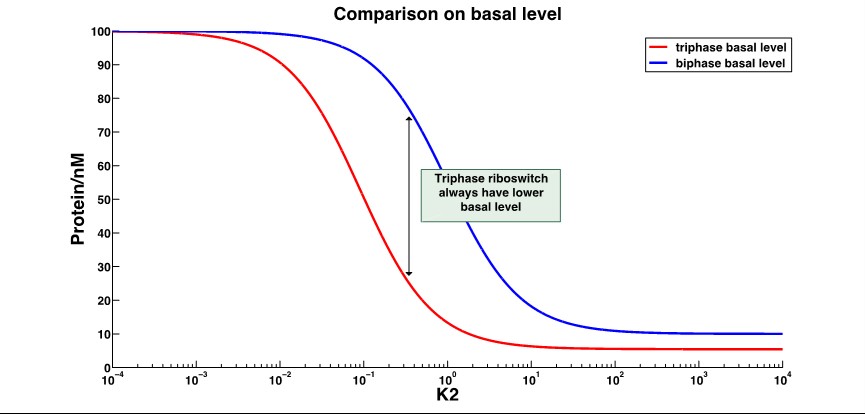
| + | '''2.Basal level expression'''(with ON behavior riboswitch): | ||
| + | [[File:AleaderT4.jpg|1000px|thumb|center| '''Figure 5'''.With the dominant conformation B, triphase riboswitch always have a lower basal level than the biphase riboswitch, with the changing k2. And the lowerlimit of triphase riboswitch is lower as well. | ||
| + | ''k2'':distribution constant between different conformations ]] | ||
| + | That shows the leakage expression of triphase riboswitch significantly decrease compared to the biphase riboswitch. | ||
| − | We tested the | + | == Manipulating and Experimental Data (Quantitative Measurement) == |
| − | [[File: | + | We tested the ALeaderT performance by measuring the fluorescence intensity of mRFP1 when different concentrations of kanamycin sulfate are added to the bacteria culture. |
| − | + | [[File:101AleaderT.jpg|600px|thumb|center| '''Figure 6''' . The dose-response curve of ALeaderT with a promoter J23101, measured by mRFP fluorescence test.]] | |
| − | + | More data? See [https://parts.igem.org/Part:BBa_K1100014 BBa_K11000014] | |
| − | [ | + | == Reference == |
| + | [1] Jia X, Zhang J, Sun W, et al. Riboswitch Control of Aminoglycoside Antibiotic Resistance[J]. Cell, 2013, 152(1): 68-81. | ||
| − | [[ | + | [2] Hanau‐Berçot B, Podglajen I, Casin I, et al. An intrinsic control element for translational initiation in class 1 integrons[J]. Molecular microbiology, 2002, 44(1): 119-130. |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Latest revision as of 01:30, 28 September 2013
J23101-ALeaderT
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Background
Riboswitches are regulatory RNAs that regulate gene expression by binding small molecules. Recently, a novel riboswitch was reported which is present in the leader RNA of the aminoglycoside resistance genes that encode the aminoglycoside acetyl transferase (AAC) and aminoglycoside adenyl transferase (AAD) enzymes. When the aminoglycosides bind to the leader RNA, a conformational change will be induced, leading to the expression of the aminoglycoside resistance genes (Jia X et al, 2013.). In addition, the leader RNA encompass an integrase site (attl1), which is overlapped by a short peptide encoded by an ORF (ORF11) (Berçot B, et al, 2002.). Therefore, Aleader is a complicated riboswitch containing ORF11, cistron, aminoglycoside-binding domain and att1 recombination site. Due to the complicacy of Aleader, the secondary structure of it could not be predicted by most software such as Mfold and NUPACK (Jia X et al, 2013.).
As shown in Figure 1, the 75nt RNA sequence of Aleader contains two SD sequences (ribosome binding sites) and an anti-SD sequence (CUUC) which can complementarily pair with either of the SD sequences. In the absence of aminoglycosides, anti-SD pairs with SD2. The binding of ribosomes to SD1 triggers the translation of a small peptide which stops at the stop codon ahead of SD2, and therefore inhibits the translation of the desired gene after SD2. When aminoglycosides (kanamycin for example) exists, it will induce a structural change of Aleader. The anti-SD sequence base-pairs with SD1, consequently unmasking SD2 for ribosomal binding, which results in the translation of the following gene.
Our design of AleaderT
Many riboswitches have lower dynamic range and higher leakage rate compared with protein regulators. To reduce the basal level and enhance the dynamic range, while having no effect on the protein product, we designed a novel riboswitch called ALeaderT, following the principle of three-phase switch design.
We added a GGGS linker (dock III), a common neutral peptide linker, after the SD2 and ATG. Impressively, by RNA 2D-prediction, we found that dock III will capture antiSD2 sequence to form a stable stem loop, with characteristics of terminator, though neutral as an amino peptide. We suppose that the ALeaderT will have three different steady-state conformations, A “Translation off Termination off”, B “Translation on Termination off”, and C “Translation on Termination on”(Figure 3).
The A conformation is the ligand-free conformation that SD2 and antiSD form the stem loop, resulting in no translation nor termination. The B conformation is the ligand-binding conformation that SD2 is released and SD1 binds the antiSD, leading to the translation. And in C conformation, the SD2 is also released, making it possible to bind the ribosome, but the antiSD and dock III form a stable stem loop to reduce the transcription. Through mathematical modeling, we found that the leakage expression would significantly decrease, while the dynamic range increased several-fold. The working range is usually changed, as well.
1.Dynamic Range(shown with ratio):
2.Basal level expression(with ON behavior riboswitch):
That shows the leakage expression of triphase riboswitch significantly decrease compared to the biphase riboswitch.
Manipulating and Experimental Data (Quantitative Measurement)
We tested the ALeaderT performance by measuring the fluorescence intensity of mRFP1 when different concentrations of kanamycin sulfate are added to the bacteria culture.
More data? See BBa_K11000014
Reference
[1] Jia X, Zhang J, Sun W, et al. Riboswitch Control of Aminoglycoside Antibiotic Resistance[J]. Cell, 2013, 152(1): 68-81.
[2] Hanau‐Berçot B, Podglajen I, Casin I, et al. An intrinsic control element for translational initiation in class 1 integrons[J]. Molecular microbiology, 2002, 44(1): 119-130.