Difference between revisions of "Part:BBa K1051200"
| (3 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
<br /> | <br /> | ||
<h3>Principle</h3> | <h3>Principle</h3> | ||
| − | + | ||
| − | + | ||
| − | + | ||
http://upload.wikimedia.org/wikipedia/commons/thumb/5/5b/Ubiquitylation.svg/800px-Ubiquitylation.svg.png | http://upload.wikimedia.org/wikipedia/commons/thumb/5/5b/Ubiquitylation.svg/800px-Ubiquitylation.svg.png | ||
| + | <br /> | ||
The ubiquitination system. | The ubiquitination system. | ||
| + | <br /> | ||
| + | '''Ubiquitin''',a small regulatory protein that has been found in almost all tissues of eukaryotic organisms,Ubiquitination is a post translational modification where ubuiquitin is attached to a substrate protein.It is carried out in three main steps; activation, conjugation and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s).<br /> | ||
| + | We want to install ubiquitin to fluorescent proteins at N-terminal.<br /> | ||
| + | To test the function of ubiquitin fusion protein, we construct measurement pathway K1051288, which contains J63006, K1051200,K1051003 and K1051006. | ||
https://static.igem.org/mediawiki/2013/6/68/Growth-Curve.jpg | https://static.igem.org/mediawiki/2013/6/68/Growth-Curve.jpg | ||
| + | <br /> | ||
Yeast growth curve. | Yeast growth curve. | ||
| + | https://static.igem.org/mediawiki/2013/3/3f/Sc_bright_20.jpg | ||
| + | <br /> | ||
| + | We made the chip as a platform for watching and synchronize the cells. First step is capture the cells by the chip. As showed in the figure, budding yeast can be captured by the chip successfully. | ||
<!-- Add more about the biology of this part her | <!-- Add more about the biology of this part her | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| Line 25: | Line 32: | ||
<partinfo>BBa_K1051200 parameters</partinfo> | <partinfo>BBa_K1051200 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | <h3>References</h3> | ||
| + | [1] Wilkinson KD (October 2005). "The discovery of ubiquitin-dependent proteolysis". Proc. Natl. Acad. Sci. U.S.A. 102 (43): 15280–2. doi:10.1073/pnas.0504842102. PMC 1266097. PMID 16230621. | ||
| + | <br /> | ||
| + | [2] Kimura Y, Tanaka K (2010). "Regulatory mechanisms involved in the control of ubiquitin homeostasis". J Biochem 147 (6): 793–8. doi:10.1093/jb/mvq044. PMID 20418328. | ||
Latest revision as of 18:32, 27 September 2013
Ubiquitin
Purpose
Works for the degradation of fluorescent proteins we used.
Principle
http://upload.wikimedia.org/wikipedia/commons/thumb/5/5b/Ubiquitylation.svg/800px-Ubiquitylation.svg.png
The ubiquitination system.
Ubiquitin,a small regulatory protein that has been found in almost all tissues of eukaryotic organisms,Ubiquitination is a post translational modification where ubuiquitin is attached to a substrate protein.It is carried out in three main steps; activation, conjugation and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s).
We want to install ubiquitin to fluorescent proteins at N-terminal.
To test the function of ubiquitin fusion protein, we construct measurement pathway K1051288, which contains J63006, K1051200,K1051003 and K1051006.
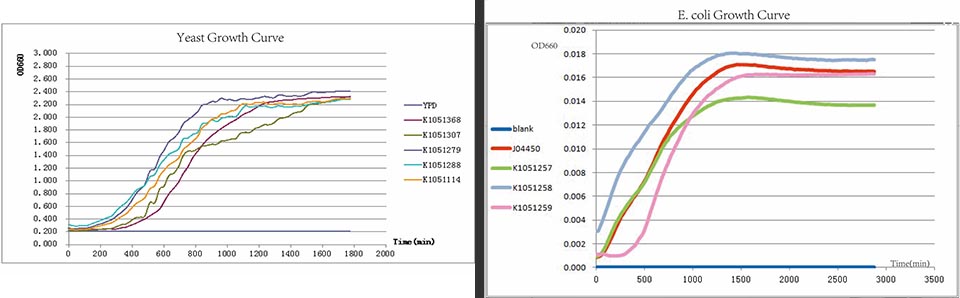
Yeast growth curve.

We made the chip as a platform for watching and synchronize the cells. First step is capture the cells by the chip. As showed in the figure, budding yeast can be captured by the chip successfully.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 233
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 25
Illegal AgeI site found at 253 - 1000COMPATIBLE WITH RFC[1000]
References
[1] Wilkinson KD (October 2005). "The discovery of ubiquitin-dependent proteolysis". Proc. Natl. Acad. Sci. U.S.A. 102 (43): 15280–2. doi:10.1073/pnas.0504842102. PMC 1266097. PMID 16230621.
[2] Kimura Y, Tanaka K (2010). "Regulatory mechanisms involved in the control of ubiquitin homeostasis". J Biochem 147 (6): 793–8. doi:10.1093/jb/mvq044. PMID 20418328.
