Difference between revisions of "Part:BBa K1119009"
(→Result) |
|||
| (2 intermediate revisions by one other user not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1119009 short</partinfo> | <partinfo>BBa_K1119009 short</partinfo> | ||
| − | In characterization of Mitochondrial Leader Sequence ([[Part:BBa_K1119001|BBa_K1119001]]), the CDS of MLS was assembled in frame with that of GFP reporter using Freiburg’s RFC25 format([[Part:BBa_K648013|BBa_K648013]]). The translation unit was driven by CMV promoter ([[Part:BBa_K1119006|BBa_K1119006]]) and terminated by hGH polyA signal ([[Part:BBa_K404108|BBa_K404108]]). This construct was used for characterization of MLS BioBrick([[Part:BBa_K1119001|BBa_K1119001]]) . | + | In characterization of Mitochondrial Leader Sequence(MLS) ([[Part:BBa_K1119001|BBa_K1119001]]), the CDS of MLS was assembled in frame with that of GFP reporter using Freiburg’s RFC25 format([[Part:BBa_K648013|BBa_K648013]]). The translation unit was driven by CMV promoter ([[Part:BBa_K1119006|BBa_K1119006]]) and terminated by hGH polyA signal ([[Part:BBa_K404108|BBa_K404108]]). This construct was used for characterization of MLS BioBrick([[Part:BBa_K1119001|BBa_K1119001]]) . |
== Result == | == Result == | ||
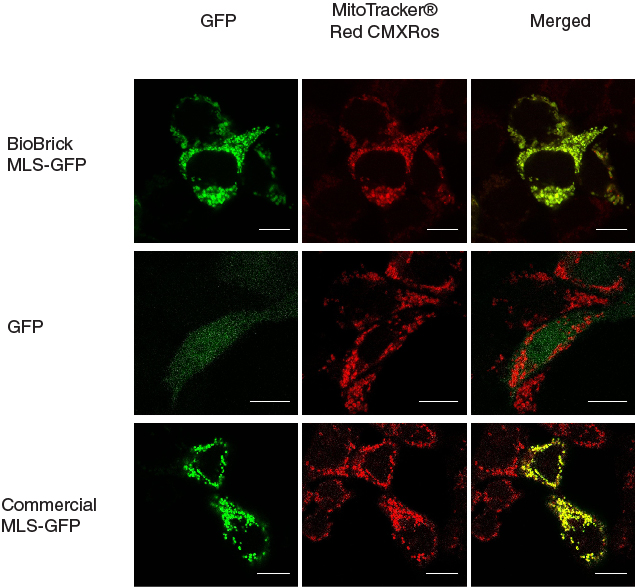
| − | This MLS-GFP generator (BBa_K1119009) was transfected into HEK293FT cells. Mitochondria were stained after transfection and co-localization was determined by area of signal that overlapped. | + | This MLS-GFP generator ([[Part:BBa_K1119009|BBa_K1119009]]) was transfected into HEK293FT cells. Mitochondria were stained after transfection and co-localization was determined by area of signal that overlapped. |
To provide a positive control, CDS of EGFP from pEGFP-N1 (Clontech) was inserted downstream and in frame with the CDS of the MLS in the commercial plasmid pCMV/myc/mito, (Invitrogen, Carlsbard, CA). A negative control was made by GFP generator that does not contains the CDS of MLS (BBa_K1119008). | To provide a positive control, CDS of EGFP from pEGFP-N1 (Clontech) was inserted downstream and in frame with the CDS of the MLS in the commercial plasmid pCMV/myc/mito, (Invitrogen, Carlsbard, CA). A negative control was made by GFP generator that does not contains the CDS of MLS (BBa_K1119008). | ||
| − | [[File:mlschar_1.jpg|600px|thumb|center|'''Figure 1. MLS directs GFP into mitochondria.''' When MLS is added to the N terminus of GFP, the GFP was directed to the mitochondria in the cells, giving patches of GFP signal that overlapped with the signals from MitoTracker®. When MLS is not added to the GFP, the GFP signal can be seen scattered all around in the cell. | + | [[File:mlschar_1.jpg|600px|thumb|center|'''Figure 1. MLS directs GFP into mitochondria.''' When MLS is added to the N terminus of GFP, the GFP was directed to the mitochondria in the cells, giving patches of GFP signal that overlapped with the signals from MitoTracker®. When MLS is not added to the GFP, the GFP signal can be seen scattered all around in the cell. Scale bar = 10 microns]] |
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 15:36, 26 September 2013
CMV promoter - MLS - GFP - hGH polyA tail
In characterization of Mitochondrial Leader Sequence(MLS) (BBa_K1119001), the CDS of MLS was assembled in frame with that of GFP reporter using Freiburg’s RFC25 format(BBa_K648013). The translation unit was driven by CMV promoter (BBa_K1119006) and terminated by hGH polyA signal (BBa_K404108). This construct was used for characterization of MLS BioBrick(BBa_K1119001) .
Result
This MLS-GFP generator (BBa_K1119009) was transfected into HEK293FT cells. Mitochondria were stained after transfection and co-localization was determined by area of signal that overlapped.
To provide a positive control, CDS of EGFP from pEGFP-N1 (Clontech) was inserted downstream and in frame with the CDS of the MLS in the commercial plasmid pCMV/myc/mito, (Invitrogen, Carlsbard, CA). A negative control was made by GFP generator that does not contains the CDS of MLS (BBa_K1119008).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 614
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 628
Illegal AgeI site found at 1462 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1865
Illegal BsaI.rc site found at 1367
