Difference between revisions of "Part:BBa K639003:Experience"
(→User Reviews) |
|||
| (15 intermediate revisions by 2 users not shown) | |||
| Line 11: | Line 11: | ||
|- | |- | ||
|width='10%'| | |width='10%'| | ||
| − | <partinfo>BBa_K639003 AddReview | + | <partinfo>BBa_K639003 AddReview Number</partinfo> |
| − | <I> | + | <I>Team name</I> |
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
| − | + | Review text | |
| − | |} | + | |} |
| + | |||
| + | |||
<!-- End of the user review template --> | <!-- End of the user review template --> | ||
<!-- DON'T DELETE --><partinfo>BBa_K639003 EndReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_K639003 EndReviews</partinfo> | ||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K639003 AddReview 3</partinfo> | ||
| + | <I>Imperial College London iGEM 2013</I> | ||
| + | |width='60%' valign='top'| | ||
| + | Stress biosensor expresses mCherry in "non stress" conditions. However we did characterise and utilise this part in several of our experiments. | ||
| + | |} | ||
<h2>Imperial iGEM 2013</h2> | <h2>Imperial iGEM 2013</h2> | ||
| − | + | <h3>Stress biosensor characterisation (BBa_K639003)</h3> | |
| − | + | Originally we intended on using [https://parts.igem.org/Part:BBa_K639003 BBa_K639003] to detect whether our cells were stressed when grown with an array of potentially toxic plastics and degradation products. However, as our data below shows, this biobrick is very leaky. As an alternative we utilised the stress sensor as a marker for our chassis E. coli (MG1655) for an array of qualitative and quantitative waste growth and toxicity assays. | |
| + | <b>Note:</b> The stress sensor induces mCherry production through a mechanism involving the ppGpp stress response. Induction with IPTG bypassess this mechanism through an inhibition of LacI, resulting in mCherry expression. | ||
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| − | | [[File:OD_stress.png|thumbnail|right|400px| | + | |[[File:OD_stress.png|thumbnail|right|400px|<b>Stress sensor growth assay</b>E. coli (MG1655) transformed with stress biosensor (BBa_K639003) and grown at 37oC, with shaking for 6 hours. Error bars represent S.E.M. n=4 ]] |
| − | | [[File: | + | |[[File:Fluorescence_stress.png|thumbnail|right|400px|<b>Stress sensor IPTG induced fluorescence</b>E. coli (MG1655) transformed with stress biosensor (BBa_K639003) and grown at 37oC, with shaking for 6 hours. Error bars represent S.E.M. n=4]] |
| + | |- | ||
| + | |||
| + | |[[File:StressResponse.jpg|400px|thumb|left|<b>BBa_K639003 transformed into E coli. strain MG1655.</b> Pink colonies are visible, which relate to 'leaky' mCherry production]] | ||
| + | |||
| + | |[[File:K639003IPTG.jpeg|500px|thumb|left|<b>BBa_K639003 transformed into E coli. strain MG1655.</b> Cells were grown at 37oC in 4ml LB with 0, 1 or 2mM IPTG. At 6 hours post IPTG induction, cells were spun down and imaged.]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | <h3>Long term waste growth assays</h3> | ||
| + | |||
| + | These assays were designed to test whether our chassis, E. coli (MG1655) could grow directly with waste over a long period of time. Details about waste media and waste conditioned media can be found here [http://2013.igem.org/Team:Imperial_College/Protocols Imperial 2013 protocols] | ||
| + | |||
| + | <h3>Waste media </h3> | ||
| + | |||
| + | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| + | |[[File:Waste_cocktail.png|thumbnail|center|400px|<b>LB-waste assay</b><b>(A)</b> Waste media<b>(B)</b> Ecoli containing mCherry stress biosensor (BBa_K639003) were grown in mixed waste <b>(A)</b> over 3 days, then streaked in a qualitative assay to check for growth. <b>(C)</b> mCherry stress biosensor (BBa_K639003) transformed Ecoli were streaked again after 7 days growth in SRF.]] | ||
| + | |[[File:PBS_Plus_Waste.jpg|thumbnail|center|400px|<b>PBS-waste assay</b><b>(A)</b> waste media made up in PBS (phosphate buffered saline). <b>(B)</b> E coli expressing mCherry stress biosensor (BBa_K639003) grown in waste media <b>(A)</b> over 3 days, then streaked onto an antibiotic containing plate to qualitatively assess whether the E. coli had survived. <b>(C)</b> Streaked again after 6 days growth in SRF.]] | ||
| + | |} | ||
| + | |||
| + | <b>Conclusion: MG1655 E. coli are viable and grow on mixed waste alone. Therefore we have established that our chassis could survive in a mixed waste bio-reactor context, which is validation of our concept to industrially implement our system.</b> | ||
| + | |||
| + | |||
| + | <h3>Waste conditioned media</h3> | ||
| + | |||
| + | These assays were designed to test whether our chassis, E. coli (MG1655) could grow with waste conditioned media (WCM) over a period of 24-48 hours. Waste conditioned media is a filter sterilised version of the waste media and was designed for several reasons; Firstly we were unsure whether mixed waste would be toxic to Ecoli and hence a less concentrated version may be more suitable and secondly large chunks of waste would prevent accurate OD600 measurements and therefore we decided to filter out the largest chunks. | ||
| + | |||
| + | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| + | |[[File:WCM_media.png|thumbnail|right|400px|<b>Growth assay in waste conditioned media (WCM)</b>. E. coli (MG1655) transformed with [https://parts.igem.org/Part:BBa_K639003 mCherry stress biosensor.] were grown with LB-based waste conditioned media at 37ºC, with shaking. Error bars represent S.E.M., n=4]] | ||
| + | |||
| + | |[[File:Very_stressed_ecoli.jpg|thumbnail|left|400px|<b>Photograph of waste conditioned media cultures</b> mCherry stress biosensor (BBa_K639003) transformed MG1655 were grown with [http://2013.igem.org/Team:Imperial_College/Protocols LB-WCM] at 37oC, with shaking for 48 hours.]] | ||
| + | |||
| + | [[File:WCM2.png|thumbnail|center|400px|<b>Growth assay in waste conditioned media (WCM).</b> E. coli (MG1655) transformed with either empty vector control (EV) or mCherry stress biosensor (BBa_K639003) were grown in WCM for 5 hours, with shaking at 37oC. Error bars are SEM, n=4.]] | ||
| + | |||
|} | |} | ||
| − | |||
| − | + | <b> Conclusion: MG1655 transformed with either empty vector (EV) control or mCherry stress biosensor (BBa_K639003) vector are viable and can grow in waste conditioned media. Therefore waste conditioned media is an appropriate and novel experimental media with which to characterise biobricks within a mixed waste/landfill context. These data are also characterisation of an existing biobrick (BBa_K639003) </b> | |
| − | + | <h3>Painting</h3> | |
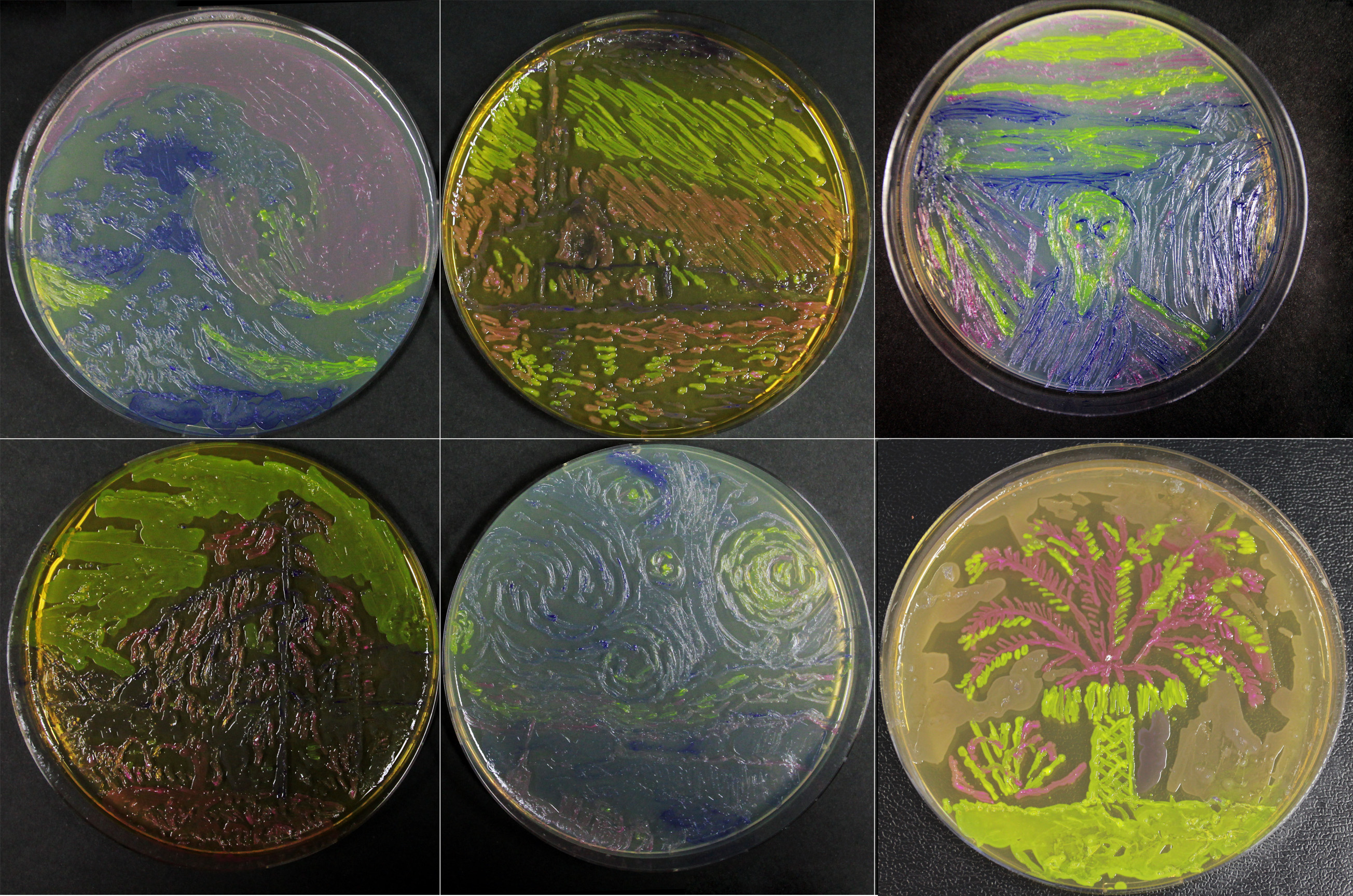
| − | [[File: | + | In addition to this, we have shown that bacterial paintings are achievable with this amazing biobrick (red): |
| + | [[File:Originals_combined_small.jpg|600px]] | ||
Latest revision as of 00:25, 1 October 2013
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K639003
User Reviews
UNIQ3df803a77f11248f-partinfo-00000000-QINU UNIQ3df803a77f11248f-partinfo-00000001-QINU
|
•••
Imperial College London iGEM 2013 |
Stress biosensor expresses mCherry in "non stress" conditions. However we did characterise and utilise this part in several of our experiments. |
Imperial iGEM 2013
Stress biosensor characterisation (BBa_K639003)
Originally we intended on using BBa_K639003 to detect whether our cells were stressed when grown with an array of potentially toxic plastics and degradation products. However, as our data below shows, this biobrick is very leaky. As an alternative we utilised the stress sensor as a marker for our chassis E. coli (MG1655) for an array of qualitative and quantitative waste growth and toxicity assays.
Note: The stress sensor induces mCherry production through a mechanism involving the ppGpp stress response. Induction with IPTG bypassess this mechanism through an inhibition of LacI, resulting in mCherry expression.
Long term waste growth assays
These assays were designed to test whether our chassis, E. coli (MG1655) could grow directly with waste over a long period of time. Details about waste media and waste conditioned media can be found here [http://2013.igem.org/Team:Imperial_College/Protocols Imperial 2013 protocols]
Waste media
Conclusion: MG1655 E. coli are viable and grow on mixed waste alone. Therefore we have established that our chassis could survive in a mixed waste bio-reactor context, which is validation of our concept to industrially implement our system.
Waste conditioned media
These assays were designed to test whether our chassis, E. coli (MG1655) could grow with waste conditioned media (WCM) over a period of 24-48 hours. Waste conditioned media is a filter sterilised version of the waste media and was designed for several reasons; Firstly we were unsure whether mixed waste would be toxic to Ecoli and hence a less concentrated version may be more suitable and secondly large chunks of waste would prevent accurate OD600 measurements and therefore we decided to filter out the largest chunks.
 Growth assay in waste conditioned media (WCM). E. coli (MG1655) transformed with mCherry stress biosensor. were grown with LB-based waste conditioned media at 37ºC, with shaking. Error bars represent S.E.M., n=4 |
Conclusion: MG1655 transformed with either empty vector (EV) control or mCherry stress biosensor (BBa_K639003) vector are viable and can grow in waste conditioned media. Therefore waste conditioned media is an appropriate and novel experimental media with which to characterise biobricks within a mixed waste/landfill context. These data are also characterisation of an existing biobrick (BBa_K639003)
Painting
In addition to this, we have shown that bacterial paintings are achievable with this amazing biobrick (red):