Difference between revisions of "Part:BBa K1062002:Design"
(→Sources) |
|||
| (5 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
| − | == | + | =='''Design Notes'''== |
| − | + | ||
| + | Part:BBa_K1062002 is a gRNA that specifically targets the middle of the XylE gene. | ||
| + | ==='''gRNA'''=== | ||
| + | A gRNA is a 102 nt sequence in which contains: a 20 nt sequence which will contain the sequence of the intended target, a 42 nt Cas9-binding hairping, and a 40 nt transcription terminator. | ||
| − | + | [[File:gRNA with NNN sequence.png]] | |
| − | + | '''Figure 1: An image show a general picture of a gRNA.''' | |
| − | === | + | |
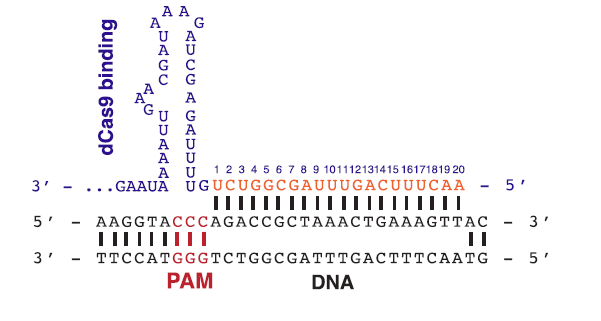
| + | An important thing to note when using or creating a gRNA is that before the 20nt sequence, '''''there must be a 3nt sequence called a PAM sequence which contain this sequence 5'-NGG-3''''''. This 3nt sequence is what the Cas9 use as a binding signal. It is also important to point out that the PAM sequence is never in the gRNA itself. | ||
| + | |||
| + | [[File:pam.png]] | ||
| + | |||
| + | '''Figure 2: An example of a PAM sequence before the 20nt target sequence.''' | ||
| + | ==Sources== | ||
| + | The gRNA was design by the iGEM team by using APE file. Once the gRNA was finalized, the team ordered primers and used PCR sewing in order to create the gRNA. | ||
| + | |||
| + | ==References== | ||
Latest revision as of 20:44, 17 September 2013
Guide RNA (gRNA) target for XylE (middle of gene)
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Design Notes
Part:BBa_K1062002 is a gRNA that specifically targets the middle of the XylE gene.
gRNA
A gRNA is a 102 nt sequence in which contains: a 20 nt sequence which will contain the sequence of the intended target, a 42 nt Cas9-binding hairping, and a 40 nt transcription terminator.
Figure 1: An image show a general picture of a gRNA.
An important thing to note when using or creating a gRNA is that before the 20nt sequence, there must be a 3nt sequence called a PAM sequence which contain this sequence 5'-NGG-3'. This 3nt sequence is what the Cas9 use as a binding signal. It is also important to point out that the PAM sequence is never in the gRNA itself.
Figure 2: An example of a PAM sequence before the 20nt target sequence.
Sources
The gRNA was design by the iGEM team by using APE file. Once the gRNA was finalized, the team ordered primers and used PCR sewing in order to create the gRNA.