Difference between revisions of "Part:BBa K1077003"
MikeFerguson (Talk | contribs) |
|||
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1077003 short</partinfo> | <partinfo>BBa_K1077003 short</partinfo> | ||
| − | J23100 fim switch b0034 GFP | + | J23100 fim switch b0034 GFP ON oreintation |
| + | |||
| + | [[File:michiganigempartmainfig1.jpg]] | ||
| + | |||
| + | Fig. 1 - The fim transcriptor is capable of changing states completely and unidirectionally | ||
| + | |||
| + | NEB 10-beta E. coli, which lack the native fim switch (fimS) and known fim recombinases, were co-transformed with either constitutive hbiF or fimE and BBa_K1077007 (J23100 fim switch, ON orientation), plated on LB plates and grown overnight for ~16 hours. The state of the switch was assayed by using an asymmetric digest assay on PCR amplified switch. There are two hincII sites located within the K1077007 switch, one of which changes position depending on the state of the switch. The result is that when the switch is in the ON position, a 870bp, 273bp, and 248bp band is produced. When the switch is in the OFF position, a 680bp, 473bp, and 273bp band is produced. A and B)The digest assay was quantified using densitometry and showed greater than 95% of the switch in the desired state (ON when co transformed with constitutive hbiF and OFF when co transformed with constitutive fimE). This is consistent with hbiF’s previously observed functionality of catalyzing the inversion of fimS from the OFF to ON orientation and fime’s previously observed functionality of catalyzing the inversion of fimS from the ON to OFF orientation. C and D) A gel of the digest assay fragments quantified in A and B. | ||
| + | |||
| + | The faint ~870bp band in 1D, seemingly indicating residual OFF state, may correspond to the constitutive recombinase generator which is 858bp. We did not have time to cure the bacteria of the constitutive recombinase generator plasmid. Additionally, the switch is on the high copy pSB1C3 plasmid and so it could be that some switch plasmids are escaping recombination. We did not have time to move the switch to a low copy plasmid or the chromosome. | ||
| + | |||
| + | [[File:michiganigempartmainfig2.jpg]] | ||
| + | |||
| + | Fig 2. - An inducible fim transcriptor system changes states and produces protein output | ||
| + | |||
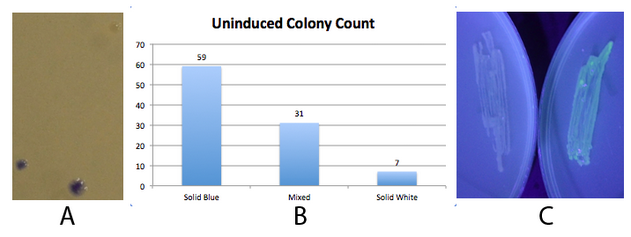
| + | NEB 10-beta E. coli, which lack the native fim switch (fimS) and known fim recombinases, were co-transformed with K1077007(amilCP j23100 fim switch) or K1077003(GFP j23100 fim switch), and K1077002 (aTc inducible fimE, HSL inducible hbiF) and plated on to LB plates with or without inducer. A) Close up of 3 colonies on a plate containing K1077007 and K1077002 co transformants. Three distinct phenotypes were observed: Solid Blue colonies(bottom left), Mixed colonies that had distinct white and blue regions(bottom middle), and Solid White colonies(top right). Therefore, the uninduced fim transcriptor in the context of K1077002 is subject to leaky fimE and/or hbiF activity. B) Quantification of the phenotypes observed on the plate in A. C) When induced with 4.32µM aTc and grown overnight (left) , no GFP is produced as expected due to induced fimE turning the switch OFF. When induced with 1µM HSL and grown overnight (right), GFP is produced as expected due to induced hbiF turning the switch ON. | ||
| + | |||
| + | Given the absence of GFP in the aTc induced switch shown in 2C, leaky hbiF expression is overcome by the induced fimE expression. Nothing can be said about induced hbiF overcoming leaky fimE since the green may still indicate only some of the plasmids are flipped. Whether or not the phenotypes of the colonies observed in 2A are due to variable leakage rates of each recombinase or variable enzymatic activity of each of the recombinases, or both, can not be determined. Since these are co transformations, the white and mixed colonies observed could simply be due to chance production of fimE only or first in the first generation, flipping the whole population of switch plasmids in the cell (~1). Subsequent generations might inherit the switch mostly in the initial state. | ||
| + | We did generate only inducible hbiF and fimE plasmids, but did not have time to co transform them with the switch or submit them to the registry. These would allow us to test the effects of leakage of each recombinase on the switch. Other assays, such as time course data of recombinase activity under the same inducible system, would be needed to determine relative rates of recombinase activity. Moving the inducible recombinase generator to a low copy plasmid, may reduce leakage effects. Ideally, a more tightly regulated inducible system should be coupled to the recombinases. Promoter and/or rbs optimization should be performed to minimize recombinase leakage above the threshold needed for significant flipping activity. | ||
| + | |||
| + | |||
| + | Sources: | ||
| + | |||
| + | 1. Schwan WR. Regulation of fim genes in uropathogenic Escherichia coli. World J Clin Infect Dis 2011; 1(1): 1725. | ||
| + | |||
| + | 2. I. C. Blomfield, D. H. Kulasekara and B. I. Eisenstein. Integration host factor stimulates both FimB- and FimE-mediated site-specific DNA inversion that controls phase variation of type 1 fimbriae expression in Escherichia coli. Molecular Microbiology (1997) 23(4), 705–717. | ||
| + | |||
| + | 3. M. P. McCusker, E. C. Turner and C. J. Dorman. DNA sequence heterogeneity in Fim tyrosine-integrase recombinase-binding elements and functional motif asymmetries determine the directionality of the fim genetic switch in Escherichia coli K-12. Molecular Microbiology, 67, 171–187. | ||
| + | |||
| + | 4. Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996 Dec 27;87(7):1295-306. | ||
| + | |||
| + | 5. Wang Q, Calvo JM. Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of a higher-order nucleoprotein structure. EMBO J. 1993 Jun;12(6):2495-501. | ||
| + | |||
| + | 6. Jerome Bonnet, Pakpoom Subsoontorn, and Drew Endy. Rewritable digital data storage in live cells via engineered control of recombination directionality. PNAS. 2012 Apr 6. | ||
| + | |||
| + | 7. D. L. Gally, J. Leathart and I. C. Blomfield. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Molecular Microbiology (1996) 21(4), 725–738. | ||
| + | |||
| + | 8. Ham et al. A Tightly Regulated Inducible Expression System Utilizing the fim Inversion Recombination Switch. Biotechnology and Bioengineering, Vol. 94, No. 1, May 5, 2006. | ||
| + | |||
| + | 9. Jerome Bonnet et al. Amplifying Genetic Logic Gates. Science 3 May 2013, Vol. 340 no. 6132 pp. 599-603. | ||
| + | |||
| + | 10. Ham TS, Lee SK, Keasling JD, Arkin AP. Design and Construction of a Double Inversion Recombination Switch for Heritable Sequential Genetic Memory. PLoS ONE, 2008, 3(7): e2815. doi:10.1371/journal.pone.0002815 | ||
| + | |||
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | ||
| + | Fim System Background | ||
| + | |||
| + | The natural fim system from E. coli utilizes temperature sensitive phase variation, through enzyme-catalyzed inversion of a promoter containing segment of DNA, as a means to control production of fimbriae. The fimA promoter is the promoter being flipped. When it is transcribing the fimA gene, the state of switch is said to be in the ON phase. When it is facing the reverse direction on the complementary strand, the switch is said to be in the OFF phase. The primary known components of the fim system are 4 site specific tyrosine recombinases fimE, fimB, hbiF/ipbA, and ipuA; the fim switch (fimS); and the fimS regulators leucine-responsive regulatory protein (LRP) and Integration Host Factor (IHF). | ||
| + | |||
| + | It is worth mentioning the directionality of each of the 4 recombinases in flipping the switch. FimE flips the switch unidirectionally from phase ON to OFF. HbiF flips the switch unidirectionally from OFF to ON. FimB flips both ON to OFF and OFF to ON with a slight preference for the OFF to ON direction. IpuA was shown to flip the switch unidirectionally from OFF to ON. | ||
| + | |||
| + | In addition to LRP and IHF which act to regulate the actual inversion event, possibly through an involved DNA bending protein complex (fig 2), there exists the OFF to ON inhibitory protein papB and a host of other hypothetical and known regulators acting either directly on fimS, fimA, or the recombinase genes and their regulators. For a complete review on the fim system, refer to [1]. | ||
| + | |||
| + | [[File:michiganpartbackgroundfig1.jpg]] | ||
| + | |||
| + | Fig 1: Schematic of fim system and regulators. Source: [1] | ||
| + | |||
| + | [[File:michiganpartbackgroundfig2.jpg]] | ||
| + | |||
| + | Fig 2: Theoretical model of the association of regulators with the fim switch. Source: [2] | ||
| + | |||
| + | |||
| + | |||
| + | Fim Switch (fimS) Background | ||
| + | |||
| + | The fimA promoter lies in between two 9bp segments of DNA referred to as inverted repeats (IRs); an inverted repeat left (IRL) and an inverted repeat right (IRR). When flipping the fimS switch, the recombinases bind directly to these inverted repeats and create a break in the DNA. Upon inversion, the IRs are restored to their same sequences. The IRR is identical to the reverse complement of the IRL and vice versa, hence the name. The sequences immediately adjacent to either side of the 9bp IRs are part of the recombinase binding sites and are referred to as half-sites. Since the 9bp IRs is where the DNA is broken, one of the half site’s sequence is part of the actual segment of DNA being flipped and is variable depending on the switch’s state. These are the internal half sites. The external half sites never change and are used to identify the IRs as either inverted repeat left (IRL) or inverted repeat right (IRR). The proteins fimB and fimE have been shown to bind to these inverted repeats by DNA footprinting and are necessary to catalyze inversion of the approximately 314bp fimS DNA in between the IR’s. Though not formally shown, it is very likely that the 2 other known recombinases hbiF and ipuA also bind to these IRs. | ||
| + | |||
| + | [[File:michiganpartbackgroundfig3.jpg]] | ||
| + | |||
| + | Fig 3: Diagram showing the ON and OFF state sequences of the inverted repeats. Source: [3] | ||
| + | |||
| + | The IHF and LRP are DNA bending proteins that have been shown to bind to the fimS DNA. Both proteins are very well characterized ([4],[5]). IHF bends DNA by 160 degrees. It is a necessary component of the Int/Ex system. LRP bends DNA by 52 degrees upon binding to a single LRP binding site. When LRP binds to two adjacent LRP binding sites, the angle of bending can be increased to at least 135 degrees. Within and immediately upstream of the fimS DNA, there are two IHF binding sites (IHFI, II) and three LRP binding sites (LRPI, II, III). See fig 3 for their relative location. Although the exact mechanism of fimS inversion is unknown, it is likely that both IHF and LRP are critical components of the inversion event, especially given their radical effects on the DNA. | ||
| + | |||
| + | As shown by the 2012 Michigan igem team, inversion is extremely rare on a switch containing only the IRL and IRR. Whether or not this is due to the lack of interaction with IHF and LRP or due to the effects of negative regulators can not be determined. In vitro assays in which purified IHF, LRP, and recombinase proteins are mixed with a fim switch need to be performed. If successful in vitro, the system may be more readily ported to other species. | ||
| + | |||
| + | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K1077003 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1077003 SequenceAndFeatures</partinfo> | ||
Latest revision as of 15:57, 29 September 2013
J23100 fim switch b0034 GFP
J23100 fim switch b0034 GFP ON oreintation
Fig. 1 - The fim transcriptor is capable of changing states completely and unidirectionally
NEB 10-beta E. coli, which lack the native fim switch (fimS) and known fim recombinases, were co-transformed with either constitutive hbiF or fimE and BBa_K1077007 (J23100 fim switch, ON orientation), plated on LB plates and grown overnight for ~16 hours. The state of the switch was assayed by using an asymmetric digest assay on PCR amplified switch. There are two hincII sites located within the K1077007 switch, one of which changes position depending on the state of the switch. The result is that when the switch is in the ON position, a 870bp, 273bp, and 248bp band is produced. When the switch is in the OFF position, a 680bp, 473bp, and 273bp band is produced. A and B)The digest assay was quantified using densitometry and showed greater than 95% of the switch in the desired state (ON when co transformed with constitutive hbiF and OFF when co transformed with constitutive fimE). This is consistent with hbiF’s previously observed functionality of catalyzing the inversion of fimS from the OFF to ON orientation and fime’s previously observed functionality of catalyzing the inversion of fimS from the ON to OFF orientation. C and D) A gel of the digest assay fragments quantified in A and B.
The faint ~870bp band in 1D, seemingly indicating residual OFF state, may correspond to the constitutive recombinase generator which is 858bp. We did not have time to cure the bacteria of the constitutive recombinase generator plasmid. Additionally, the switch is on the high copy pSB1C3 plasmid and so it could be that some switch plasmids are escaping recombination. We did not have time to move the switch to a low copy plasmid or the chromosome.
Fig 2. - An inducible fim transcriptor system changes states and produces protein output
NEB 10-beta E. coli, which lack the native fim switch (fimS) and known fim recombinases, were co-transformed with K1077007(amilCP j23100 fim switch) or K1077003(GFP j23100 fim switch), and K1077002 (aTc inducible fimE, HSL inducible hbiF) and plated on to LB plates with or without inducer. A) Close up of 3 colonies on a plate containing K1077007 and K1077002 co transformants. Three distinct phenotypes were observed: Solid Blue colonies(bottom left), Mixed colonies that had distinct white and blue regions(bottom middle), and Solid White colonies(top right). Therefore, the uninduced fim transcriptor in the context of K1077002 is subject to leaky fimE and/or hbiF activity. B) Quantification of the phenotypes observed on the plate in A. C) When induced with 4.32µM aTc and grown overnight (left) , no GFP is produced as expected due to induced fimE turning the switch OFF. When induced with 1µM HSL and grown overnight (right), GFP is produced as expected due to induced hbiF turning the switch ON.
Given the absence of GFP in the aTc induced switch shown in 2C, leaky hbiF expression is overcome by the induced fimE expression. Nothing can be said about induced hbiF overcoming leaky fimE since the green may still indicate only some of the plasmids are flipped. Whether or not the phenotypes of the colonies observed in 2A are due to variable leakage rates of each recombinase or variable enzymatic activity of each of the recombinases, or both, can not be determined. Since these are co transformations, the white and mixed colonies observed could simply be due to chance production of fimE only or first in the first generation, flipping the whole population of switch plasmids in the cell (~1). Subsequent generations might inherit the switch mostly in the initial state. We did generate only inducible hbiF and fimE plasmids, but did not have time to co transform them with the switch or submit them to the registry. These would allow us to test the effects of leakage of each recombinase on the switch. Other assays, such as time course data of recombinase activity under the same inducible system, would be needed to determine relative rates of recombinase activity. Moving the inducible recombinase generator to a low copy plasmid, may reduce leakage effects. Ideally, a more tightly regulated inducible system should be coupled to the recombinases. Promoter and/or rbs optimization should be performed to minimize recombinase leakage above the threshold needed for significant flipping activity.
Sources:
1. Schwan WR. Regulation of fim genes in uropathogenic Escherichia coli. World J Clin Infect Dis 2011; 1(1): 1725.
2. I. C. Blomfield, D. H. Kulasekara and B. I. Eisenstein. Integration host factor stimulates both FimB- and FimE-mediated site-specific DNA inversion that controls phase variation of type 1 fimbriae expression in Escherichia coli. Molecular Microbiology (1997) 23(4), 705–717.
3. M. P. McCusker, E. C. Turner and C. J. Dorman. DNA sequence heterogeneity in Fim tyrosine-integrase recombinase-binding elements and functional motif asymmetries determine the directionality of the fim genetic switch in Escherichia coli K-12. Molecular Microbiology, 67, 171–187.
4. Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996 Dec 27;87(7):1295-306.
5. Wang Q, Calvo JM. Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of a higher-order nucleoprotein structure. EMBO J. 1993 Jun;12(6):2495-501.
6. Jerome Bonnet, Pakpoom Subsoontorn, and Drew Endy. Rewritable digital data storage in live cells via engineered control of recombination directionality. PNAS. 2012 Apr 6.
7. D. L. Gally, J. Leathart and I. C. Blomfield. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Molecular Microbiology (1996) 21(4), 725–738.
8. Ham et al. A Tightly Regulated Inducible Expression System Utilizing the fim Inversion Recombination Switch. Biotechnology and Bioengineering, Vol. 94, No. 1, May 5, 2006.
9. Jerome Bonnet et al. Amplifying Genetic Logic Gates. Science 3 May 2013, Vol. 340 no. 6132 pp. 599-603.
10. Ham TS, Lee SK, Keasling JD, Arkin AP. Design and Construction of a Double Inversion Recombination Switch for Heritable Sequential Genetic Memory. PLoS ONE, 2008, 3(7): e2815. doi:10.1371/journal.pone.0002815
Usage and Biology
Fim System Background
The natural fim system from E. coli utilizes temperature sensitive phase variation, through enzyme-catalyzed inversion of a promoter containing segment of DNA, as a means to control production of fimbriae. The fimA promoter is the promoter being flipped. When it is transcribing the fimA gene, the state of switch is said to be in the ON phase. When it is facing the reverse direction on the complementary strand, the switch is said to be in the OFF phase. The primary known components of the fim system are 4 site specific tyrosine recombinases fimE, fimB, hbiF/ipbA, and ipuA; the fim switch (fimS); and the fimS regulators leucine-responsive regulatory protein (LRP) and Integration Host Factor (IHF).
It is worth mentioning the directionality of each of the 4 recombinases in flipping the switch. FimE flips the switch unidirectionally from phase ON to OFF. HbiF flips the switch unidirectionally from OFF to ON. FimB flips both ON to OFF and OFF to ON with a slight preference for the OFF to ON direction. IpuA was shown to flip the switch unidirectionally from OFF to ON.
In addition to LRP and IHF which act to regulate the actual inversion event, possibly through an involved DNA bending protein complex (fig 2), there exists the OFF to ON inhibitory protein papB and a host of other hypothetical and known regulators acting either directly on fimS, fimA, or the recombinase genes and their regulators. For a complete review on the fim system, refer to [1].
Fig 1: Schematic of fim system and regulators. Source: [1]
Fig 2: Theoretical model of the association of regulators with the fim switch. Source: [2]
Fim Switch (fimS) Background
The fimA promoter lies in between two 9bp segments of DNA referred to as inverted repeats (IRs); an inverted repeat left (IRL) and an inverted repeat right (IRR). When flipping the fimS switch, the recombinases bind directly to these inverted repeats and create a break in the DNA. Upon inversion, the IRs are restored to their same sequences. The IRR is identical to the reverse complement of the IRL and vice versa, hence the name. The sequences immediately adjacent to either side of the 9bp IRs are part of the recombinase binding sites and are referred to as half-sites. Since the 9bp IRs is where the DNA is broken, one of the half site’s sequence is part of the actual segment of DNA being flipped and is variable depending on the switch’s state. These are the internal half sites. The external half sites never change and are used to identify the IRs as either inverted repeat left (IRL) or inverted repeat right (IRR). The proteins fimB and fimE have been shown to bind to these inverted repeats by DNA footprinting and are necessary to catalyze inversion of the approximately 314bp fimS DNA in between the IR’s. Though not formally shown, it is very likely that the 2 other known recombinases hbiF and ipuA also bind to these IRs.
Fig 3: Diagram showing the ON and OFF state sequences of the inverted repeats. Source: [3]
The IHF and LRP are DNA bending proteins that have been shown to bind to the fimS DNA. Both proteins are very well characterized ([4],[5]). IHF bends DNA by 160 degrees. It is a necessary component of the Int/Ex system. LRP bends DNA by 52 degrees upon binding to a single LRP binding site. When LRP binds to two adjacent LRP binding sites, the angle of bending can be increased to at least 135 degrees. Within and immediately upstream of the fimS DNA, there are two IHF binding sites (IHFI, II) and three LRP binding sites (LRPI, II, III). See fig 3 for their relative location. Although the exact mechanism of fimS inversion is unknown, it is likely that both IHF and LRP are critical components of the inversion event, especially given their radical effects on the DNA.
As shown by the 2012 Michigan igem team, inversion is extremely rare on a switch containing only the IRL and IRR. Whether or not this is due to the lack of interaction with IHF and LRP or due to the effects of negative regulators can not be determined. In vitro assays in which purified IHF, LRP, and recombinase proteins are mixed with a fim switch need to be performed. If successful in vitro, the system may be more readily ported to other species.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 306
Illegal NheI site found at 329 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1049